The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide
- PMID: 20676057
- PMCID: PMC2944061
- DOI: 10.1038/emboj.2010.180
The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide
Abstract
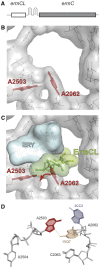
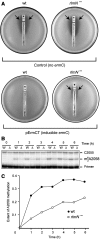
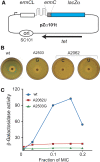
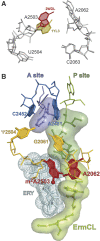
The ribosome is able to monitor the structure of the nascent peptide and can stall in response to specific peptide sequences. Such programmed stalling is used for the regulation of gene expression. The molecular mechanisms of the nascent-peptide recognition and ribosome stalling are unknown. We identified the conserved and posttranscriptionally modified 23S rRNA nucleotide m(2)A2503 located at the entrance of the ribosome exit tunnel as a key component of the ribosomal response mechanism. A2503 mutations abolish nascent-peptide-dependent stalling at the leader cistrons of several inducible antibiotic resistance genes and at the secM regulatory gene. Remarkably, lack of the C2 methylation of A2503 significantly function induction of expression of the ermC gene, indicating that the functional role of posttranscriptional modification is to fine-tune ribosome-nascent peptide interactions. Structural and biochemical evidence suggest that m(2)A2503 may act in concert with the previously identified nascent-peptide sensor, A2062, in the ribosome exit tunnel to relay the stalling signal to the peptidyl transferase centre.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

