The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models
- PMID: 20676074
- PMCID: PMC3315840
- DOI: 10.1038/nbt.1665
The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models
Abstract
Gene expression data from microarrays are being applied to predict preclinical and clinical endpoints, but the reliability of these predictions has not been established. In the MAQC-II project, 36 independent teams analyzed six microarray data sets to generate predictive models for classifying a sample with respect to one of 13 endpoints indicative of lung or liver toxicity in rodents, or of breast cancer, multiple myeloma or neuroblastoma in humans. In total, >30,000 models were built using many combinations of analytical methods. The teams generated predictive models without knowing the biological meaning of some of the endpoints and, to mimic clinical reality, tested the models on data that had not been used for training. We found that model performance depended largely on the endpoint and team proficiency and that different approaches generated models of similar performance. The conclusions and recommendations from MAQC-II should be useful for regulatory agencies, study committees and independent investigators that evaluate methods for global gene expression analysis.
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at
Figures
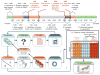
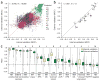
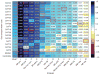
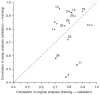
Comment in
-
Consistency of predictive signature genes and classifiers.Pharmacogenomics. 2011 Apr;12(4):461-3. doi: 10.2217/pgs.11.26. Pharmacogenomics. 2011. PMID: 21521018 No abstract available.
References
-
- Marshall E. Getting the noise out of gene arrays. Science. 2004;306:630–631. - PubMed
-
- Frantz S. An array of problems. Nat Rev Drug Discov. 2005;4:362–363. - PubMed
-
- Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. - PubMed
-
- Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet. 2003;362:1439–1444. - PubMed
-
- Ioannidis JP. Microarrays and molecular research: noise discovery? Lancet. 2005;365:454–455. - PubMed
Publication types
MeSH terms
Grants and funding
- CA55819-01A1/CA/NCI NIH HHS/United States
- R01 RR021967/RR/NCRR NIH HHS/United States
- Z99 MH999999/ImNIH/Intramural NIH HHS/United States
- R01 GM083226/GM/NIGMS NIH HHS/United States
- CA97513-01/CA/NCI NIH HHS/United States
- R00 CA126184/CA/NCI NIH HHS/United States
- T32GM074906/GM/NIGMS NIH HHS/United States
- R33 CA097513/CA/NCI NIH HHS/United States
- 1R01GM083084-01/GM/NIGMS NIH HHS/United States
- R01 GM072611/GM/NIGMS NIH HHS/United States
- 1R01RR021967-01A2/RR/NCRR NIH HHS/United States
- T32 GM074906/GM/NIGMS NIH HHS/United States
- R01 GM083084/GM/NIGMS NIH HHS/United States
- Z99 CL999999/ImNIH/Intramural NIH HHS/United States
- Z99 HL999999/ImNIH/Intramural NIH HHS/United States
- Z99 LM999999/ImNIH/Intramural NIH HHS/United States
- P01 CA055819/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

