Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin
- PMID: 20676099
- PMCID: PMC2930131
- DOI: 10.1038/ng.630
Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin
Abstract
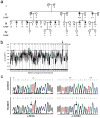
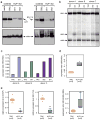
Hereditary persistence of fetal hemoglobin (HPFH) is characterized by persistent high levels of fetal hemoglobin (HbF) in adults. Several contributory factors, both genetic and environmental, have been identified but others remain elusive. HPFH was found in 10 of 27 members from a Maltese family. We used a genome-wide SNP scan followed by linkage analysis to identify a candidate region on chromosome 19p13.12-13. Sequencing revealed a nonsense mutation in the KLF1 gene, p.K288X, which ablated the DNA-binding domain of this key erythroid transcriptional regulator. Only family members with HPFH were heterozygous carriers of this mutation. Expression profiling on primary erythroid progenitors showed that KLF1 target genes were downregulated in samples from individuals with HPFH. Functional assays suggested that, in addition to its established role in regulating adult globin expression, KLF1 is a key activator of the BCL11A gene, which encodes a suppressor of HbF expression. These observations provide a rationale for the effects of KLF1 haploinsufficiency on HbF levels.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


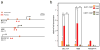
Comment in
-
Putting a finger on the switch.Nat Genet. 2010 Sep;42(9):733-4. doi: 10.1038/ng0910-733. Nat Genet. 2010. PMID: 20802474 Free PMC article.
References
-
- Serjeant G. Geographic heterogeneity of sickle cell disease. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of Hemoglobin. Cambridge University Press; Cambrige: 2001. pp. 895–906.
-
- Bieker JJ. Probing the onset and regulation of erythroid cell-specific gene expression. Mt Sinai J Med. 2005;72:333–8. - PubMed
-
- Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–42. - PubMed
-
- Stamatoyannopoulos G, Grosveld F. Hemoglobin switching. In: Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H, editors. The Molecular Basis of Blood Diseases. WB Saunders Company; Philadelphia PA: 2001. pp. 135–182.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

