G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae
- PMID: 20676380
- PMCID: PMC2908698
- DOI: 10.1371/journal.pcbi.1000861
G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae
Abstract
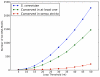
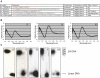
G-quadruplex DNA is a four-stranded DNA structure formed by non-Watson-Crick base pairing between stacked sets of four guanines. Many possible functions have been proposed for this structure, but its in vivo role in the cell is still largely unresolved. We carried out a genome-wide survey of the evolutionary conservation of regions with the potential to form G-quadruplex DNA structures (G4 DNA motifs) across seven yeast species. We found that G4 DNA motifs were significantly more conserved than expected by chance, and the nucleotide-level conservation patterns suggested that the motif conservation was the result of the formation of G4 DNA structures. We characterized the association of conserved and non-conserved G4 DNA motifs in Saccharomyces cerevisiae with more than 40 known genome features and gene classes. Our comprehensive, integrated evolutionary and functional analysis confirmed the previously observed associations of G4 DNA motifs with promoter regions and the rDNA, and it identified several previously unrecognized associations of G4 DNA motifs with genomic features, such as mitotic and meiotic double-strand break sites (DSBs). Conserved G4 DNA motifs maintained strong associations with promoters and the rDNA, but not with DSBs. We also performed the first analysis of G4 DNA motifs in the mitochondria, and surprisingly found a tenfold higher concentration of the motifs in the AT-rich yeast mitochondrial DNA than in nuclear DNA. The evolutionary conservation of the G4 DNA motif and its association with specific genome features supports the hypothesis that G4 DNA has in vivo functions that are under evolutionary constraint.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
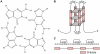



References
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. - PubMed
-
- Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

