Comprehensive transcriptome analysis of mouse embryonic stem cell adipogenesis unravels new processes of adipocyte development
- PMID: 20678241
- PMCID: PMC2945782
- DOI: 10.1186/gb-2010-11-8-r80
Comprehensive transcriptome analysis of mouse embryonic stem cell adipogenesis unravels new processes of adipocyte development
Abstract
Background: The current epidemic of obesity has caused a surge of interest in the study of adipose tissue formation. While major progress has been made in defining the molecular networks that control adipocyte terminal differentiation, the early steps of adipocyte development and the embryonic origin of this lineage remain largely unknown.
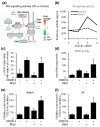
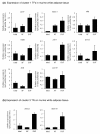
Results: Here we performed genome-wide analysis of gene expression during adipogenesis of mouse embryonic stem cells (ESCs). We then pursued comprehensive bioinformatic analyses, including de novo functional annotation and curation of the generated data within the context of biological pathways, to uncover novel biological functions associated with the early steps of adipocyte development. By combining in-depth gene regulation studies and in silico analysis of transcription factor binding site enrichment, we also provide insights into the transcriptional networks that might govern these early steps.
Conclusions: This study supports several biological findings: firstly, adipocyte development in mouse ESCs is coupled to blood vessel morphogenesis and neural development, just as it is during mouse development. Secondly, the early steps of adipocyte formation involve major changes in signaling and transcriptional networks. A large proportion of the transcription factors that we uncovered in mouse ESCs are also expressed in the mouse embryonic mesenchyme and in adipose tissues, demonstrating the power of our approach to probe for genes associated with early developmental processes on a genome-wide scale. Finally, we reveal a plethora of novel candidate genes for adipocyte development and present a unique resource that can be further explored in functional assays.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

