Bat coronaviruses and experimental infection of bats, the Philippines
- PMID: 20678314
- PMCID: PMC3298303
- DOI: 10.3201/eid1608.100208
Bat coronaviruses and experimental infection of bats, the Philippines
Abstract
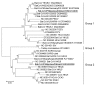
Fifty-two bats captured during July 2008 in the Philippines were tested by reverse transcription-PCR to detect bat coronavirus (CoV) RNA. The overall prevalence of virus RNA was 55.8%. We found 2 groups of sequences that belonged to group 1 (genus Alphacoronavirus) and group 2 (genus Betacoronavirus) CoVs. Phylogenetic analysis of the RNA-dependent RNA polymerase gene showed that groups 1 and 2 CoVs were similar to Bat-CoV/China/A515/2005 (95% nt sequence identity) and Bat-CoV/HKU9-1/China/2007 (83% identity), respectively. To propagate group 2 CoVs obtained from a lesser dog-faced fruit bat (Cynopterus brachyotis), we administered intestine samples orally to Leschenault rousette bats (Rousettus leschenaulti) maintained in our laboratory. After virus replication in the bats was confirmed, an additional passage of the virus was made in Leschenault rousette bats, and bat pathogenesis was investigated. Fruit bats infected with virus did not show clinical signs of infection.
Figures

Similar articles
-
Detection and phylogenetic analysis of group 1 coronaviruses in South American bats.Emerg Infect Dis. 2008 Dec;14(12):1890-3. doi: 10.3201/eid1412.080642. Emerg Infect Dis. 2008. PMID: 19046513 Free PMC article.
-
Group B betacoronavirus in rhinolophid bats, Japan.J Vet Med Sci. 2014 Sep;76(9):1267-9. doi: 10.1292/jvms.14-0012. Epub 2014 May 27. J Vet Med Sci. 2014. PMID: 24871548 Free PMC article.
-
Neotropical Bats from Costa Rica harbour Diverse Coronaviruses.Zoonoses Public Health. 2015 Nov;62(7):501-5. doi: 10.1111/zph.12181. Epub 2015 Feb 4. Zoonoses Public Health. 2015. PMID: 25653111 Free PMC article.
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
-
Coronaviruses in Bats: A Review for the Americas.Viruses. 2021 Jun 25;13(7):1226. doi: 10.3390/v13071226. Viruses. 2021. PMID: 34201926 Free PMC article. Review.
Cited by
-
Molecular Detection and Genetic Characterization of Novel RNA Viruses in Wild and Synanthropic Rodents and Shrews in Kenya.Front Microbiol. 2019 Nov 21;10:2696. doi: 10.3389/fmicb.2019.02696. eCollection 2019. Front Microbiol. 2019. PMID: 31824465 Free PMC article.
-
Bats, emerging infectious diseases, and the rabies paradigm revisited.Emerg Health Threats J. 2011 Jun 20;4:7159. doi: 10.3402/ehtj.v4i0.7159. Emerg Health Threats J. 2011. PMID: 24149032 Free PMC article.
-
Long-term surveillance of bat coronaviruses in Korea: Diversity and distribution pattern.Transbound Emerg Dis. 2020 Nov;67(6):2839-2848. doi: 10.1111/tbed.13653. Epub 2020 Jun 15. Transbound Emerg Dis. 2020. PMID: 32473082 Free PMC article.
-
Global patterns of phylogenetic diversity and transmission of bat coronavirus.Sci China Life Sci. 2023 Apr;66(4):861-874. doi: 10.1007/s11427-022-2221-5. Epub 2022 Nov 11. Sci China Life Sci. 2023. PMID: 36378474 Free PMC article.
-
Coronavirus surveillance in wildlife from two Congo basin countries detects RNA of multiple species circulating in bats and rodents.PLoS One. 2021 Jun 9;16(6):e0236971. doi: 10.1371/journal.pone.0236971. eCollection 2021. PLoS One. 2021. PMID: 34106949 Free PMC article.
References
-
- Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–900. 10.1128/JVI.79.18.11892-11900.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
