Transcriptional regulation by p53
- PMID: 20679336
- PMCID: PMC2908772
- DOI: 10.1101/cshperspect.a000935
Transcriptional regulation by p53
Abstract
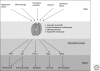
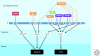
Inactivation of p53 is critical for the formation of most tumors. Illumination of the key function(s) of p53 protein in protecting cells from becoming cancerous is therefore a worthy goal. Arguably p53's most important function is to act as a transcription factor that directly regulates perhaps several hundred of the cell's RNA polymerase II (RNAP II)-transcribed genes, and indirectly regulates thousands of others. Indeed p53 is the most well studied mammalian transcription factor. The p53 tetramer binds to its response element where it can recruit diverse transcriptional coregulators such as histone modifying enzymes, chromatin remodeling factors, subunits of the mediator complex, and components of general transcription machinery and preinitiation complex (PIC) to modulate RNAPII activity at target loci (Laptenko and Prives 2006). The p53 transcriptional program is regulated in a stimulus-specific fashion (Murray-Zmijewski et al. 2008; Vousden and Prives 2009), whereby distinct subsets of p53 target genes are induced in response to different p53-activating agents, likely allowing cells to tailor their response to different types of stress. How p53 is able to discriminate between these different loci is the subject of intense research. Here, we describe key aspects of the fundamentals of p53-mediated transcriptional regulation and target gene promoter selectivity.
Figures


References
-
- An W, Kim J, Roeder RG 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748 - PubMed
-
- Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J 2005. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem 280: 26776–26787 - PubMed
-
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89: 1175–1184 - PubMed
-
- Balakrishnan SK, Gross DS 2008. The tumor suppressor p53 associates with gene coding regions and co-traverses with elongating RNA polymerase II in an in vivo model. Oncogene 27: 2661–2672 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
