Demonstrating polymorphic miRNA-mediated gene regulation in vivo: application to the g+6223G->A mutation of Texel sheep
- PMID: 20679369
- PMCID: PMC2924544
- DOI: 10.1261/rna.2131110
Demonstrating polymorphic miRNA-mediated gene regulation in vivo: application to the g+6223G->A mutation of Texel sheep
Abstract
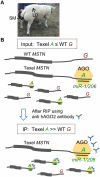
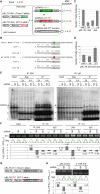
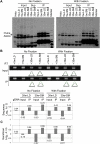
We herein describe the development of a biochemical method to evaluate the effect of single nucleotide polymorphisms (SNPs) in target genes on their regulation by microRNAs in vivo. The method is based on the detection of allelic imbalance in RNAs coimmunoprecipitated with AGO proteins from tissues of heterozygous individuals. We characterize the performances of our approach using a model system in a cell culture, and then apply it successfully to prove that the 3'UTR g+6223G-->A mutation operates by promoting RISC-dependent down-regulation of myostatin (MSTN) in skeletal muscle of Texel sheep.
Figures

References
-
- Abelson J, Kwan K, O'Roak B, Baek D, Stillman A, Morgan T, Mathews C, Pauls D, Rasin M, Gunel M, et al. 2005. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science 310: 317–320 - PubMed
-
- Adams B, Furneaux H, White B 2007. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21: 1132–1147 - PubMed
-
- Beitzinger M, Peters L, Zhu J, Kremmer E, Meister G 2007. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol 4: 76–84 - PubMed
-
- Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, Hemminki K, Lenner P, Forsti A 2008. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis 29: 1394–1399 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
