Antisense oligonucleotides and spinal muscular atrophy: skipping along
- PMID: 20679391
- PMCID: PMC2912553
- DOI: 10.1101/gad.1961710
Antisense oligonucleotides and spinal muscular atrophy: skipping along
Abstract
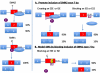
Antisense oligonucleotides (ASOs) can be used to alter the splicing of a gene and either restore production of a required protein or eliminate a toxic product. In this issue of Genes & Development, Hua and colleagues (pp. 1634-1644) show that ASOs directed against an intron splice silencer (ISS) in the survival motor neuron 2 (SMN2) gene alter the amount of full-length SMN transcript in the nervous system, restoring SMN to levels that could correct spinal muscular atrophy (SMA).
Figures
Comment on
-
Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model.Genes Dev. 2010 Aug 1;24(15):1634-44. doi: 10.1101/gad.1941310. Epub 2010 Jul 12. Genes Dev. 2010. PMID: 20624852 Free PMC article.
References
-
- Butchbach ME, Singh J, Thorsteinsdottir M, Saieva L, Slominski E, Thurmond J, Andresson T, Zhang J, Edwards JD, Simard LR, et al. 2010. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet 19: 454–467 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
