An inexpensive, specific and highly sensitive protocol to detect the BrafV600E mutation in melanoma tumor biopsies and blood
- PMID: 20679909
- PMCID: PMC2936688
- DOI: 10.1097/CMR.0b013e32833d8d48
An inexpensive, specific and highly sensitive protocol to detect the BrafV600E mutation in melanoma tumor biopsies and blood
Abstract
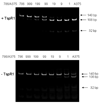
The Braf(V600E) mutation has been detected in patients with metastatic melanoma, colon, thyroid and other cancers. Recent studies suggested that tumors with this mutation are especially sensitive to Braf inhibitors, hence the need to reliably determine the Braf status of tumor specimens. The present technologies used to screen for this mutation fail to address the problems associated with infiltrating stromal and immune cells bearing wild-type Braf alleles and thus may fail to detect the presence of mutant Braf(V600E) tumors. We have developed a rapid, inexpensive method that reduces the contamination of wild-type Braf sequences from tumor biopsies. The protocol involves a series of PCR amplifications and restriction digestions that take advantage of unique features of both wild type and mutant Braf RNA at position 600. Using this protocol, mutant Braf can be detected in RNA from mixed populations with as few as 0.1% Braf(V600E) mutant cells.
Figures



Similar articles
-
Assaying for BRAF V600E in tissue and blood in melanoma.Methods Mol Biol. 2014;1102:117-36. doi: 10.1007/978-1-62703-727-3_8. Methods Mol Biol. 2014. PMID: 24258977
-
Molecular platforms utilized to detect BRAF V600E mutation in melanoma.Semin Cutan Med Surg. 2012 Dec;31(4):267-73. doi: 10.1016/j.sder.2012.07.007. Semin Cutan Med Surg. 2012. PMID: 23174497
-
Clinical implication of highly sensitive detection of the BRAF V600E mutation in fine-needle aspirations of thyroid nodules: a comparative analysis of three molecular assays in 4585 consecutive cases in a BRAF V600E mutation-prevalent area.J Clin Endocrinol Metab. 2012 Jul;97(7):2299-306. doi: 10.1210/jc.2011-3135. Epub 2012 Apr 12. J Clin Endocrinol Metab. 2012. PMID: 22500044
-
Quantitative analysis of the BRAF V600E mutation in circulating tumor-derived DNA in melanoma patients using competitive allele-specific TaqMan PCR.Int J Clin Oncol. 2016 Oct;21(5):981-988. doi: 10.1007/s10147-016-0976-y. Epub 2016 Apr 4. Int J Clin Oncol. 2016. PMID: 27041702
-
BRAF V600E mutation-specific antibody: A review.Semin Diagn Pathol. 2015 Sep;32(5):400-8. doi: 10.1053/j.semdp.2015.02.010. Epub 2015 Feb 7. Semin Diagn Pathol. 2015. PMID: 25744437 Review.
Cited by
-
The Current State of Molecular Testing in the BRAF-Mutated Melanoma Landscape.Front Mol Biosci. 2020 Jun 30;7:113. doi: 10.3389/fmolb.2020.00113. eCollection 2020. Front Mol Biosci. 2020. PMID: 32695793 Free PMC article. Review.
-
Detecting BRAF Mutations in Formalin-Fixed Melanoma: Experiences with Two State-of-the-Art Techniques.Case Rep Oncol. 2012 May;5(2):280-9. doi: 10.1159/000339300. Epub 2012 Jun 5. Case Rep Oncol. 2012. PMID: 22740817 Free PMC article.
-
Biomarkers in melanoma: where are we now?Melanoma Manag. 2014 Nov;1(2):139-150. doi: 10.2217/mmt.14.19. Epub 2014 Dec 4. Melanoma Manag. 2014. PMID: 30190819 Free PMC article. Review.
-
Plasma Circulating Tumor DNA Levels for the Monitoring of Melanoma Patients: Landscape of Available Technologies and Clinical Applications.Biomed Res Int. 2017;2017:5986129. doi: 10.1155/2017/5986129. Epub 2017 Apr 6. Biomed Res Int. 2017. PMID: 28484715 Free PMC article. Review.
-
BRAF in Melanoma: Pathogenesis, Diagnosis, Inhibition, and Resistance.J Skin Cancer. 2011;2011:423239. doi: 10.1155/2011/423239. Epub 2011 Nov 17. J Skin Cancer. 2011. PMID: 22175026 Free PMC article.
References
-
- Vandrovcova J, Lagerstedt-Robinsson K, Påhlman L, Lindblom A. Somatic BRAF-V600E mutations in familial colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2270–2273. - PubMed
-
- Cradic KW, Milosevic D, Rosenberg AM, Erickson LA, McIver B, Grebe SK. Mutant BRAF(T1799A) can be detected in the blood of papillary thyroid carcinoma patients and correlates with disease status. J Clin Endocrinol Metab. 2009;94:5001–5009. - PubMed
-
- Orru G, Coghe F, Faa G, Pillai S, Manieli C, Montaldo C, et al. Rapid multiplex real-time PCR by molecular beacons for different BRAF allele detection in papillary thyroid carcinoma. Diagn Mol Pathol. 2010;19:1–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

