hHSS1: a novel secreted factor and suppressor of glioma growth located at chromosome 19q13.33
- PMID: 20680400
- PMCID: PMC3052511
- DOI: 10.1007/s11060-010-0314-6
hHSS1: a novel secreted factor and suppressor of glioma growth located at chromosome 19q13.33
Abstract
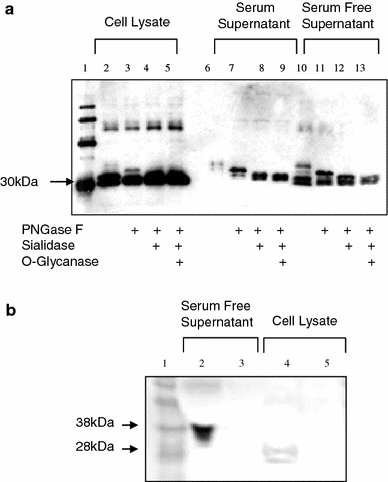
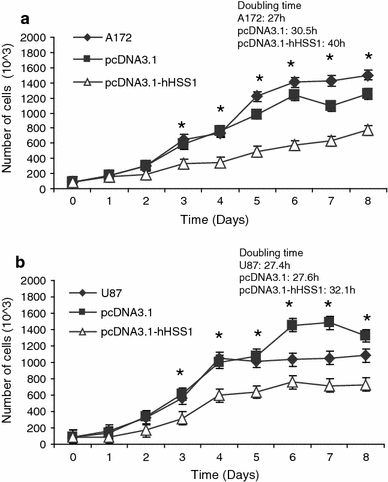
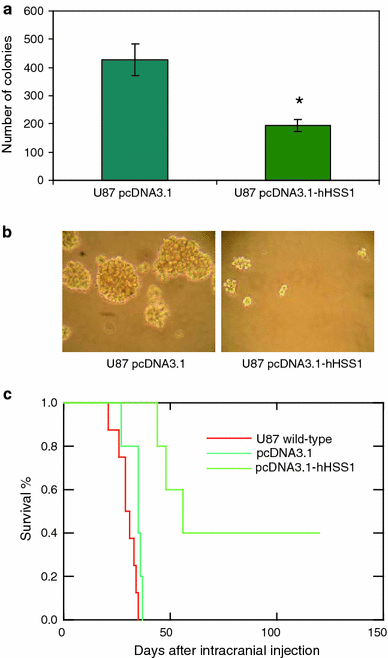
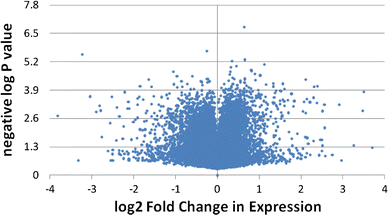
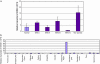
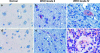
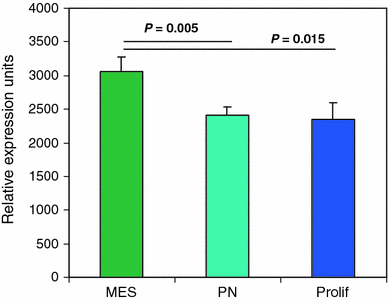
The completion of the Human Genome Project resulted in discovery of many unknown novel genes. This feat paved the way for the future development of novel therapeutics for the treatment of human disease based on novel biological functions and pathways. Towards this aim, we undertook a bioinformatics analysis of in-house microarray data derived from purified hematopoietic stem cell populations. This effort led to the discovery of HSS1 (Hematopoietic Signal peptide-containing Secreted 1) and its splice variant HSM1 (Hematopoietic Signal peptide-containing Membrane domain-containing 1). HSS1 gene is evolutionarily conserved across species, phyla and even kingdoms, including mammals, invertebrates and plants. Structural analysis showed no homology between HSS1 and known proteins or known protein domains, indicating that it was a truly novel protein. Interestingly, the human HSS1 (hHSS1) gene is located at chromosome 19q13.33, a genomic region implicated in various cancers, including malignant glioma. Stable expression of hHSS1 in glioma-derived A172 and U87 cell lines greatly reduced their proliferation rates compared to mock-transfected cells. hHSS1 expression significantly affected the malignant phenotype of U87 cells both in vitro and in vivo. Further, preliminary immunohistochemical analysis revealed an increase in hHSS1/HSM1 immunoreactivity in two out of four high-grade astrocytomas (glioblastoma multiforme, WHO IV) as compared to low expression in all four low-grade diffuse astrocytomas (WHO grade II). High-expression of hHSS1 in high-grade gliomas was further supported by microarray data, which indicated that mesenchymal subclass gliomas exclusively up-regulated hHSS1. Our data reveal that HSS1 is a truly novel protein defining a new class of secreted factors, and that it may have an important role in cancer, particularly glioma.
Figures




References
-
- Rubio M-P, Correa KM, Ueki K, et al. The putative glioma tumor suppressor gene on chromosome 19q maps between APOC2 and HRC1. Cancer Res. 1994;54:4760–4763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

