A cooperative Escherichia coli aspartate transcarbamoylase without regulatory subunits
- PMID: 20681545
- PMCID: PMC2935174
- DOI: 10.1021/bi1010333
A cooperative Escherichia coli aspartate transcarbamoylase without regulatory subunits
Abstract
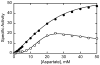
Here we report the isolation, kinetic characterization, and X-ray structure determination of a cooperative Escherichia coli aspartate transcarbamoylase (ATCase) without regulatory subunits. The native ATCase holoenzyme consists of six catalytic chains organized as two trimers bridged noncovalently by six regulatory chains organized as three dimers, c(6)r(6). Dissociation of the native holoenzyme produces catalytically active trimers, c(3), and nucleotide-binding regulatory dimers, r(2). By introducing specific disulfide bonds linking the catalytic chains from the upper trimer site specifically to their corresponding chains in the lower trimer prior to dissociation, a new catalytic unit, c(6), was isolated consisting of two catalytic trimers linked by disulfide bonds. Not only does the c(6) species display enhanced enzymatic activity compared to the wild-type enzyme, but the disulfide bonds also impart homotropic cooperativity, never observed in the wild-type c(3). The c(6) ATCase was crystallized in the presence of phosphate and its X-ray structure determined to 2.10 A resolution. The structure of c(6) ATCase liganded with phosphate exists in a nearly identical conformation as other R-state structures with similar values calculated for the vertical separation and planar angles. The disulfide bonds linking upper and lower catalytic trimers predispose the active site into a more active conformation by locking the 240s loop into the position characteristic of the high-affinity R state. Furthermore, the elimination of the structural constraints imposed by the regulatory subunits within the holoenzyme provides increased flexibility to the c(6) enzyme, enhancing its activity over the wild-type holoenzyme (c(6)r(6)) and c(3). The covalent linkage between upper and lower catalytic trimers restores homotropic cooperativity so that a binding event at one or so active sites stimulates binding at the other sites. Reduction of the disulfide bonds in the c(6) ATCase results in c(3) catalytic subunits that display kinetic parameters similar to those of wild-type c(3). This is the first report of an active c(6) catalytic unit that displays enhanced activity and homotropic cooperativity.
Figures






Similar articles
-
In vivo formation of active aspartate transcarbamoylase from complementing fragments of the catalytic polypeptide chains.Protein Sci. 1993 Jun;2(6):1013-23. doi: 10.1002/pro.5560020614. Protein Sci. 1993. PMID: 8318886 Free PMC article.
-
Three of the six possible intersubunit stabilizing interactions involving Glu-239 are sufficient for restoration of the homotropic and heterotropic properties of Escherichia coli aspartate transcarbamoylase.J Biol Chem. 2000 Jan 14;275(2):752-8. doi: 10.1074/jbc.275.2.752. J Biol Chem. 2000. PMID: 10625604
-
Reconstitution of active catalytic trimer of aspartate transcarbamoylase from proteolytically cleaved polypeptide chains.Protein Sci. 1993 Jun;2(6):1001-12. doi: 10.1002/pro.5560020613. Protein Sci. 1993. PMID: 8318885 Free PMC article.
-
Structure and mechanisms of Escherichia coli aspartate transcarbamoylase.Acc Chem Res. 2012 Mar 20;45(3):444-53. doi: 10.1021/ar200166p. Epub 2011 Oct 19. Acc Chem Res. 2012. PMID: 22011033 Free PMC article. Review.
-
From feedback inhibition to allostery: the enduring example of aspartate transcarbamoylase.FEBS J. 2014 Jan;281(2):612-20. doi: 10.1111/febs.12483. Epub 2013 Sep 5. FEBS J. 2014. PMID: 23953008 Review.
Cited by
-
Solution NMR Spectroscopy for the Study of Enzyme Allostery.Chem Rev. 2016 Jun 8;116(11):6323-69. doi: 10.1021/acs.chemrev.5b00541. Epub 2016 Jan 6. Chem Rev. 2016. PMID: 26734986 Free PMC article. Review.
-
Crystal structure and biochemical properties of putrescine carbamoyltransferase from Enterococcus faecalis: Assembly, active site, and allosteric regulation.Proteins. 2012 May;80(5):1436-47. doi: 10.1002/prot.24042. Epub 2012 Feb 13. Proteins. 2012. PMID: 22328207 Free PMC article.
-
From Genome to Structure and Back Again: A Family Portrait of the Transcarbamylases.Int J Mol Sci. 2015 Aug 12;16(8):18836-64. doi: 10.3390/ijms160818836. Int J Mol Sci. 2015. PMID: 26274952 Free PMC article. Review.
-
Allostery and cooperativity in Escherichia coli aspartate transcarbamoylase.Arch Biochem Biophys. 2012 Mar 15;519(2):81-90. doi: 10.1016/j.abb.2011.10.024. Epub 2011 Dec 16. Arch Biochem Biophys. 2012. PMID: 22198283 Free PMC article. Review.
References
-
- Gerhart JC, Pardee AB. Enzymology of control by feedback inhibition. J Biol Chem. 1962;237:891–896. - PubMed
-
- Gerhart JC, Pardee AB. The effect of the feedback Inhibitor CTP, on subunit interactions in aspartate transcarbamylase. Cold Spring Harbor Symp Quant Biol. 1963;28:491–496.
-
- Gouaux JE, Stevens RC, Lipscomb WN. Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate and CTP or ATP at 2.8 Å resolution and neutral pH. Biochemistry. 1990;29:7702–7715. - PubMed
-
- Honzatko RB, Lipscomb WN. Interactions of phosphate ligands with Escherichia coli aspartate carbamoyltransferase in the crystalline state. J Mol Biol. 1982;160:265–286. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

