Enrichment of Echinacea angustifolia with Bauer alkylamide 11 and Bauer ketone 23 increased anti-inflammatory potential through interference with cox-2 enzyme activity
- PMID: 20681645
- PMCID: PMC3738191
- DOI: 10.1021/jf1014268
Enrichment of Echinacea angustifolia with Bauer alkylamide 11 and Bauer ketone 23 increased anti-inflammatory potential through interference with cox-2 enzyme activity
Abstract
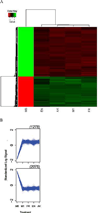
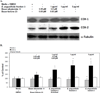
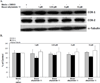
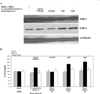
Bauer alkylamide 11 and Bauer ketone 23 were previously found to be partially responsible for Echinacea angustifolia anti-inflammatory properties. This study further tested their importance using the inhibition of prostaglandin E(2) (PGE(2)) and nitric oxide (NO) production by RAW264.7 mouse macrophages in the absence and presence of lipopolysaccharide (LPS) and E. angustifolia extracts, phytochemical enriched fractions, or pure synthesized standards. Molecular targets were probed using microarray, qRT-PCR, Western blot, and enzyme assays. Fractions with these phytochemicals were more potent inhibitors of LPS-induced PGE(2) production than E. angustifolia extracts. Microarray did not detect changes in transcripts with phytochemical treatments; however, qRT-PCR showed a decrease in TNF-alpha and an increase of iNOS transcripts. LPS-induced COX-2 protein was increased by an E. angustifolia fraction containing Bauer ketone 23 and by pure phytochemical. COX-2 activity was decreased with all treatments. The phytochemical inhibition of PGE(2) production by Echinacea may be due to the direct targeting of COX-2 enzyme.
Figures





Similar articles
-
Bauer ketones 23 and 24 from Echinacea paradoxa var. paradoxa inhibit lipopolysaccharide-induced nitric oxide, prostaglandin E2 and cytokines in RAW264.7 mouse macrophages.Phytochemistry. 2012 Feb;74:146-58. doi: 10.1016/j.phytochem.2011.10.013. Epub 2011 Nov 29. Phytochemistry. 2012. PMID: 22133644 Free PMC article.
-
Endogenous levels of Echinacea alkylamides and ketones are important contributors to the inhibition of prostaglandin E2 and nitric oxide production in cultured macrophages.J Agric Food Chem. 2009 Oct 14;57(19):8820-30. doi: 10.1021/jf901202y. J Agric Food Chem. 2009. PMID: 19807154 Free PMC article.
-
Echinacea species and alkamides inhibit prostaglandin E(2) production in RAW264.7 mouse macrophage cells.J Agric Food Chem. 2007 Sep 5;55(18):7314-22. doi: 10.1021/jf063711a. Epub 2007 Aug 15. J Agric Food Chem. 2007. PMID: 17696440 Free PMC article.
-
Echinacea increases arginase activity and has anti-inflammatory properties in RAW 264.7 macrophage cells, indicative of alternative macrophage activation.J Ethnopharmacol. 2009 Feb 25;122(1):76-85. doi: 10.1016/j.jep.2008.11.028. Epub 2008 Dec 7. J Ethnopharmacol. 2009. PMID: 19111603 Free PMC article.
-
Anti-inflammatory effect of Taraxacum officinale leaves on lipopolysaccharide-induced inflammatory responses in RAW 264.7 cells.J Med Food. 2010 Aug;13(4):870-8. doi: 10.1089/jmf.2009.1249. J Med Food. 2010. PMID: 20673058
Cited by
-
The inhibition of lipopolysaccharide-induced macrophage inflammation by 4 compounds in Hypericum perforatum extract is partially dependent on the activation of SOCS3.Phytochemistry. 2012 Apr;76:106-16. doi: 10.1016/j.phytochem.2011.12.001. Epub 2012 Jan 13. Phytochemistry. 2012. PMID: 22245632 Free PMC article.
-
ω-3 PUFA rich camelina oil by-products improve the systemic metabolism and spleen cell functions in fattening pigs.PLoS One. 2014 Oct 10;9(10):e110186. doi: 10.1371/journal.pone.0110186. eCollection 2014. PLoS One. 2014. PMID: 25303320 Free PMC article.
-
Bauer ketones 23 and 24 from Echinacea paradoxa var. paradoxa inhibit lipopolysaccharide-induced nitric oxide, prostaglandin E2 and cytokines in RAW264.7 mouse macrophages.Phytochemistry. 2012 Feb;74:146-58. doi: 10.1016/j.phytochem.2011.10.013. Epub 2011 Nov 29. Phytochemistry. 2012. PMID: 22133644 Free PMC article.
-
Echinacea purpurea (L.) Moench modulates human T-cell cytokine response.Int Immunopharmacol. 2014 Mar;19(1):94-102. doi: 10.1016/j.intimp.2013.12.019. Epub 2014 Jan 13. Int Immunopharmacol. 2014. PMID: 24434371 Free PMC article.
References
-
- Borchers AT, Keen CL, Stern JS, Gershwin ME. Inflammation and Native American medicine: the role of botanicals. Am J Clin Nutr. 2000;72(2):339–347. - PubMed
-
- Egger M, Pellett P, Nickl K, Geiger S, Graetz S, Seifert R, Heilmann J, Konig B. Synthesis and cannabinoid receptor activity of ketoalkenes from Echinacea pallida and nonnatural analogues. Chemistry. 2008;14(35):10978–10984. - PubMed
-
- Gertsch J, Schoop R, Kuenzle U, Suter A. Echinacea alkylamides modulate TNF-alpha gene expression via cannabinoid receptor CB2 and multiple signal transduction pathways. FEBS Lett. 2004;577(3):563–569. - PubMed
-
- Matthias A, Banbury L, Stevenson LM, Bone KM, Leach DN, Lehmann RP. Alkylamides from Echinacea modulate induced immune responses in macrophages. Immunol Invest. 2007;36(2):117–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

