Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift
- PMID: 20681709
- PMCID: PMC2919691
- DOI: 10.1021/ja101974k
Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift
Abstract
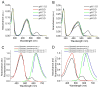
LSSmKate1 and LSSmKate2 are monomeric red fluorescent proteins (RFPs) with large Stokes shifts (LSSs), which allows for efficient separation of absorbance and emission maxima, as well as for excitation with conventional two-photon laser sources. These LSSmKates differ by a single amino acid substitution at position 160 and exhibit absorbance maxima around 460 nm, corresponding to a neutral DsRed-like chromophore. However, excitation at 460 nm leads to fluorescence emission above 600 nm. Structures of LSSmKate1 and LSSmKate2, determined at resolutions of 2.0 and 1.5 A, respectively, revealed that the predominant DsRed-chromophore configurations are cis for LSSmKate1 but trans for LSSmKate2. Crystallographic and mutagenesis analyses, as well as isotope and temperature dependences, suggest that an excited-state proton transfer (ESPT) is responsible for the LSSs observed in LSSmKates. Hydrogen bonding between the chromophore hydroxyl and Glu160 in LSSmKate1 and a proton relay involving the chromophore tyrosine hydroxyl, Ser158, and the Asp160 carboxylate in LSSmKate2 represent the putative ESPT pathways. Comparisons with mKeima LSS RFP suggest that similar proton relays could be engineered in other FPs. Accordingly, we mutated positions 158 and 160 in several conventional red-shifted FPs, including mNeptune, mCherry, mStrawberry, mOrange, and mKO, and the resulting FP variants exhibited LSS fluorescence emission in a wide range of wavelengths from 560 to 640 nm. These data suggest that different chromophores formed by distinct tripeptides in different environments can be rationally modified to yield RFPs with novel photochemical properties.
Figures



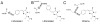


Similar articles
-
First-principles study of one- and two-photon absorption of the H-bonding complexes from monomeric red fluorescent proteins with large Stokes shifts.J Phys Chem B. 2011 Sep 15;115(36):10750-7. doi: 10.1021/jp203977m. Epub 2011 Aug 23. J Phys Chem B. 2011. PMID: 21827203
-
Characterizing the Structures, Spectra, and Energy Landscapes Involved in the Excited-State Proton Transfer Process of Red Fluorescent Protein LSSmKate1.J Phys Chem B. 2016 Sep 22;120(37):9833-42. doi: 10.1021/acs.jpcb.6b04708. Epub 2016 Sep 13. J Phys Chem B. 2016. PMID: 27581731
-
Elucidating photocycle in large Stokes shift red fluorescent proteins: Focus on mKeima.Photochem Photobiol. 2024 Jul-Aug;100(4):897-909. doi: 10.1111/php.13964. Epub 2024 May 16. Photochem Photobiol. 2024. PMID: 38752609 Review.
-
Excited state proton transfer in the red fluorescent protein mKeima.J Am Chem Soc. 2009 Sep 23;131(37):13212-3. doi: 10.1021/ja904665x. J Am Chem Soc. 2009. PMID: 19708654 Free PMC article.
-
Targeting Ultrafast Spectroscopic Insights into Red Fluorescent Proteins.Chem Asian J. 2023 Oct 17;18(20):e202300668. doi: 10.1002/asia.202300668. Epub 2023 Oct 11. Chem Asian J. 2023. PMID: 37682793 Review.
Cited by
-
Distinct effects of guanidine thiocyanate on the structure of superfolder GFP.PLoS One. 2012;7(11):e48809. doi: 10.1371/journal.pone.0048809. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23144981 Free PMC article.
-
New Red-Emitting Chloride-Sensitive Fluorescent Protein with Biological Uses.ACS Sens. 2021 Jul 23;6(7):2563-2573. doi: 10.1021/acssensors.1c00094. Epub 2021 Jun 20. ACS Sens. 2021. PMID: 34148347 Free PMC article.
-
UFObow: A single-wavelength excitable Brainbow for simultaneous multicolor ex-vivo and in-vivo imaging of mammalian cells.Commun Biol. 2024 Apr 1;7(1):394. doi: 10.1038/s42003-024-06062-3. Commun Biol. 2024. PMID: 38561421 Free PMC article.
-
Red fluorescent proteins (RFPs) and RFP-based biosensors for neuronal imaging applications.Neurophotonics. 2015 Jul;2(3):031203. doi: 10.1117/1.NPh.2.3.031203. Epub 2015 Jun 19. Neurophotonics. 2015. PMID: 26158012 Free PMC article.
-
Dissecting Optical Response and Molecular Structure of Fluorescent Proteins With Non-canonical Chromophores.Front Mol Biosci. 2020 Jul 7;7:131. doi: 10.3389/fmolb.2020.00131. eCollection 2020. Front Mol Biosci. 2020. PMID: 32733917 Free PMC article.
References
-
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Nat Methods. 2007;4:741–746. - PubMed
-
- Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A. Nat Biotechnol. 2006;24:577–581. - PubMed
-
- Violot S, Carpentier P, Blanchoin L, Bourgeois D. J Am Chem Soc. 2009;131:10356–10357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

