A sequence-based approach to identify reference genes for gene expression analysis
- PMID: 20682026
- PMCID: PMC2928167
- DOI: 10.1186/1755-8794-3-32
A sequence-based approach to identify reference genes for gene expression analysis
Abstract
Background: An important consideration when analyzing both microarray and quantitative PCR expression data is the selection of appropriate genes as endogenous controls or reference genes. This step is especially critical when identifying genes differentially expressed between datasets. Moreover, reference genes suitable in one context (e.g. lung cancer) may not be suitable in another (e.g. breast cancer). Currently, the main approach to identify reference genes involves the mining of expression microarray data for highly expressed and relatively constant transcripts across a sample set. A caveat here is the requirement for transcript normalization prior to analysis, and measurements obtained are relative, not absolute. Alternatively, as sequencing-based technologies provide digital quantitative output, absolute quantification ensues, and reference gene identification becomes more accurate.
Methods: Serial analysis of gene expression (SAGE) profiles of non-malignant and malignant lung samples were compared using a permutation test to identify the most stably expressed genes across all samples. Subsequently, the specificity of the reference genes was evaluated across multiple tissue types, their constancy of expression was assessed using quantitative RT-PCR (qPCR), and their impact on differential expression analysis of microarray data was evaluated.
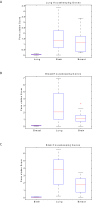
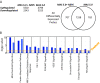
Results: We show that (i) conventional references genes such as ACTB and GAPDH are highly variable between cancerous and non-cancerous samples, (ii) reference genes identified for lung cancer do not perform well for other cancer types (breast and brain), (iii) reference genes identified through SAGE show low variability using qPCR in a different cohort of samples, and (iv) normalization of a lung cancer gene expression microarray dataset with or without our reference genes, yields different results for differential gene expression and subsequent analyses. Specifically, key established pathways in lung cancer exhibit higher statistical significance using a dataset normalized with our reference genes relative to normalization without using our reference genes.
Conclusions: Our analyses found NDUFA1, RPL19, RAB5C, and RPS18 to occupy the top ranking positions among 15 suitable reference genes optimal for normalization of lung tissue expression data. Significantly, the approach used in this study can be applied to data generated using new generation sequencing platforms for the identification of reference genes optimal within diverse contexts.
Figures

Similar articles
-
Normalization with genes encoding ribosomal proteins but not GAPDH provides an accurate quantification of gene expressions in neuronal differentiation of PC12 cells.BMC Genomics. 2010 Jan 29;11:75. doi: 10.1186/1471-2164-11-75. BMC Genomics. 2010. PMID: 20113474 Free PMC article.
-
Identification of endogenous reference genes for qRT-PCR analysis in normal matched breast tumor tissues.Oncol Res. 2009;17(8):353-65. doi: 10.3727/096504009788428460. Oncol Res. 2009. PMID: 19544972
-
Identification of new reference genes for the normalisation of canine osteoarthritic joint tissue transcripts from microarray data.BMC Mol Biol. 2007 Jul 25;8:62. doi: 10.1186/1471-2199-8-62. BMC Mol Biol. 2007. PMID: 17651481 Free PMC article.
-
Tissue-specific selection of optimal reference genes for expression analysis of anti-cancer drug-related genes in tumor samples using quantitative real-time RT-PCR.Exp Mol Pathol. 2015 Jun;98(3):375-81. doi: 10.1016/j.yexmp.2014.10.014. Epub 2014 Nov 3. Exp Mol Pathol. 2015. PMID: 25445497
-
Methods for quantitation of gene expression.Front Biosci (Landmark Ed). 2009 Jan 1;14(2):552-69. doi: 10.2741/3262. Front Biosci (Landmark Ed). 2009. PMID: 19273085 Review.
Cited by
-
Transcriptome analysis reveals global regulation in response to CO2 supplementation in oleaginous microalga Coccomyxa subellipsoidea C-169.Biotechnol Biofuels. 2016 Jul 22;9:151. doi: 10.1186/s13068-016-0571-5. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27453726 Free PMC article.
-
Cross-platform microarray meta-analysis for the mouse jejunum selects novel reference genes with highly uniform levels of expression.PLoS One. 2013 May 9;8(5):e63125. doi: 10.1371/journal.pone.0063125. Print 2013. PLoS One. 2013. PMID: 23671661 Free PMC article.
-
Differential expression of miRNAs in the serum of patients with high-risk oral lesions.Cancer Med. 2012 Oct;1(2):268-74. doi: 10.1002/cam4.17. Epub 2012 Jul 19. Cancer Med. 2012. PMID: 23342275 Free PMC article.
-
Reference genes in real-time PCR.J Appl Genet. 2013 Nov;54(4):391-406. doi: 10.1007/s13353-013-0173-x. J Appl Genet. 2013. PMID: 24078518 Free PMC article. Review.
-
Consensus reference gene(s) for gene expression studies in human cancers: end of the tunnel visible?Cell Oncol (Dordr). 2015 Dec;38(6):419-31. doi: 10.1007/s13402-015-0244-6. Epub 2015 Sep 18. Cell Oncol (Dordr). 2015. PMID: 26384826 Review.
References
-
- Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18 S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. - DOI - PubMed
-
- de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

