Herpes simplex virus UL56 interacts with and regulates the Nedd4-family ubiquitin ligase Itch
- PMID: 20682038
- PMCID: PMC2922189
- DOI: 10.1186/1743-422X-7-179
Herpes simplex virus UL56 interacts with and regulates the Nedd4-family ubiquitin ligase Itch
Abstract
Background: Herpes simplex virus type 2 (HSV-2) is one of many viruses that exploits and modifies the cellular ubiquitin system. HSV-2 expresses the tegument protein UL56 that has been implicated in cytoplasmic transport and/or release of virions, and is a putative regulatory protein of Nedd4 ubiquitin ligase. In order to elucidate the biological function of UL56, this study examined the interaction of UL56 with the Nedd4-family ubiquitin ligase Itch and its role in the regulation of Itch. Additionally, we assessed the similarity between UL56 and regulatory proteins of Itch and Nedd4, Nedd4-family-interactins proteins (Ndfip).
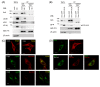
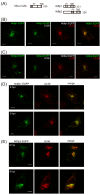
Results: UL56 interacted with Itch, independent of additional viral proteins, and mediated more striking degradation of Itch, compared to Nedd4. Moreover, it was suggested that the lysosome pathway as well as the proteasome pathway was involved in the degradation of Itch. Other HSV-2 proteins with PY motifs, such as VP5 and VP16, did not mediate the degradation of endogenous Itch. Ndfip1 and Ndfip2 were similar in subcellular distribution patterns to UL56 and colocalized with UL56 in co-transfected cells.
Conclusions: We believe that this is the first report demonstrating the interaction of a HSV-specific protein and Itch. Thus, UL56 could function as a regulatory protein of Itch. The mechanism, function and significance of regulating Itch in HSV-2 infection remain unclear and warrant further investigation.
Figures

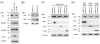


Similar articles
-
ITCH as a potential therapeutic target in human cancers.Semin Cancer Biol. 2020 Dec;67(Pt 2):117-130. doi: 10.1016/j.semcancer.2020.03.003. Epub 2020 Mar 10. Semin Cancer Biol. 2020. PMID: 32165318 Free PMC article. Review.
-
Herpes simplex virus type 2 UL56 interacts with the ubiquitin ligase Nedd4 and increases its ubiquitination.J Virol. 2008 Jun;82(11):5220-33. doi: 10.1128/JVI.02515-07. Epub 2008 Mar 19. J Virol. 2008. PMID: 18353951 Free PMC article.
-
Herpes simplex virus type 2 tegument protein UL56 relocalizes ubiquitin ligase Nedd4 and has a role in transport and/or release of virions.Virol J. 2009 Oct 16;6:168. doi: 10.1186/1743-422X-6-168. Virol J. 2009. PMID: 19835589 Free PMC article.
-
Degradation of host ubiquitin E3 ligase Itch by human cytomegalovirus UL42.J Gen Virol. 2016 Jan;97(1):196-208. doi: 10.1099/jgv.0.000336. Epub 2015 Nov 9. J Gen Virol. 2016. PMID: 26555021
-
The role of Nedd4/Nedd4-like dependant ubiquitylation in epithelial transport processes.Pflugers Arch. 2003 Jun;446(3):334-8. doi: 10.1007/s00424-003-1027-x. Epub 2003 Apr 16. Pflugers Arch. 2003. PMID: 12698368 Review.
Cited by
-
The Mouse Cytomegalovirus Gene m42 Targets Surface Expression of the Protein Tyrosine Phosphatase CD45 in Infected Macrophages.PLoS Pathog. 2016 Dec 7;12(12):e1006057. doi: 10.1371/journal.ppat.1006057. eCollection 2016 Dec. PLoS Pathog. 2016. PMID: 27926943 Free PMC article.
-
Genetic variation in NDFIP1 modifies the metabolic patterns in immune cells of multiple sclerosis patients.Sci Rep. 2021 Nov 1;11(1):21371. doi: 10.1038/s41598-021-00528-8. Sci Rep. 2021. PMID: 34725369 Free PMC article.
-
Differential microRNA profile and post-transcriptional regulation exist in systemic lupus erythematosus patients with distinct autoantibody specificities.J Clin Immunol. 2014 May;34(4):491-503. doi: 10.1007/s10875-014-0008-5. J Clin Immunol. 2014. PMID: 24659206
-
ITCH as a potential therapeutic target in human cancers.Semin Cancer Biol. 2020 Dec;67(Pt 2):117-130. doi: 10.1016/j.semcancer.2020.03.003. Epub 2020 Mar 10. Semin Cancer Biol. 2020. PMID: 32165318 Free PMC article. Review.
-
Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus?J Virol. 2019 Jun 14;93(13):e00392-19. doi: 10.1128/JVI.00392-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996099 Free PMC article. Review.
References
-
- Ball L. In: Fields Virology. 5. Knipe DM, Howley PM, editor. Vol. 1. Philadelphia: Lippincott Williams & Wilkin; 2007. Virus Replication Strategies; pp. 119–139.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

