Human genetic differentiation across the Strait of Gibraltar
- PMID: 20682051
- PMCID: PMC3020631
- DOI: 10.1186/1471-2148-10-237
Human genetic differentiation across the Strait of Gibraltar
Abstract
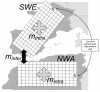
Background: The Strait of Gibraltar is a crucial area in the settlement history of modern humans because it represents a possible connection between Africa and Europe. So far, genetic data were inconclusive about the fact that this strait constitutes a barrier to gene flow, as previous results were highly variable depending on the genetic locus studied. The present study evaluates the impact of the Gibraltar region in reducing gene flow between populations from North-Western Africa and South-Western Europe, by comparing formally various genetic loci. First, we compute several statistics of population differentiation. Then, we use an original simulation approach in order to infer the most probable evolutionary scenario for the settlement of the area, taking into account the effects of both demography and natural selection at some loci.
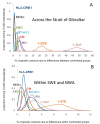
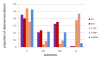
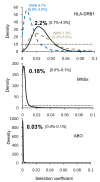
Results: We show that the genetic patterns observed today in the region of the Strait of Gibraltar may reflect an ancient population genetic structure which has not been completely erased by more recent events such as Neolithic migrations. Moreover, the differences observed among the loci (i.e. a strong genetic boundary revealed by the Y-chromosome polymorphism and, at the other extreme, no genetic differentiation revealed by HLA-DRB1 variation) across the strait suggest specific evolutionary histories like sex-mediated migration and natural selection. By considering a model of balancing selection for HLA-DRB1, we here estimate a coefficient of selection of 2.2% for this locus (although weaker in Europe than in Africa), which is in line with what was estimated from synonymous versus non-synonymous substitution rates. Selection at this marker thus appears strong enough to leave a signature not only at the DNA level, but also at the population level where drift and migration processes were certainly relevant.
Conclusions: Our multi-loci approach using both descriptive analyses and Bayesian inferences lead to better characterize the role of the Strait of Gibraltar in the evolution of modern humans. We show that gene flow across the Strait of Gibraltar occurred at relatively high rates since pre-Neolithic times and that natural selection and sex-bias migrations distorted the demographic signal at some specific loci of our genome.
Figures

Similar articles
-
Evaluating the Neolithic Expansion at Both Shores of the Mediterranean Sea.Mol Biol Evol. 2017 Dec 1;34(12):3232-3242. doi: 10.1093/molbev/msx256. Mol Biol Evol. 2017. PMID: 29029191
-
African diversity from the HLA point of view: influence of genetic drift, geography, linguistics, and natural selection.Hum Immunol. 2001 Sep;62(9):937-48. doi: 10.1016/s0198-8859(01)00293-2. Hum Immunol. 2001. PMID: 11543896
-
Y-chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa.Hum Biol. 2002 Oct;74(5):645-58. doi: 10.1353/hub.2002.0054. Hum Biol. 2002. PMID: 12495079
-
Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break?Mol Ecol. 2007 Nov;16(21):4426-44. doi: 10.1111/j.1365-294X.2007.03477.x. Epub 2007 Oct 1. Mol Ecol. 2007. PMID: 17908222 Review.
-
Conservation Status of Killer Whales, Orcinus orca, in the Strait of Gibraltar.Adv Mar Biol. 2016;75:141-172. doi: 10.1016/bs.amb.2016.07.001. Epub 2016 Aug 25. Adv Mar Biol. 2016. PMID: 27770983 Review.
Cited by
-
Complex genetic patterns in human arise from a simple range-expansion model over continental landmasses.PLoS One. 2018 Feb 21;13(2):e0192460. doi: 10.1371/journal.pone.0192460. eCollection 2018. PLoS One. 2018. PMID: 29466398 Free PMC article.
-
Spatially explicit paleogenomic simulations support cohabitation with limited admixture between Bronze Age Central European populations.Commun Biol. 2021 Oct 7;4(1):1163. doi: 10.1038/s42003-021-02670-5. Commun Biol. 2021. PMID: 34621003 Free PMC article.
-
Using uniformat and gene[rate] to Analyze Data with Ambiguities in Population Genetics.Evol Bioinform Online. 2016 Feb 21;11(Suppl 2):19-26. doi: 10.4137/EBO.S32415. eCollection 2015. Evol Bioinform Online. 2016. PMID: 26917942 Free PMC article.
-
Computer simulation of human leukocyte antigen genes supports two main routes of colonization by human populations in East Asia.BMC Evol Biol. 2015 Nov 4;15:240. doi: 10.1186/s12862-015-0512-0. BMC Evol Biol. 2015. PMID: 26530905 Free PMC article.
-
The distribution of mitochondrial DNA haplogroup H in southern Iberia indicates ancient human genetic exchanges along the western edge of the Mediterranean.BMC Genet. 2017 May 19;18(1):46. doi: 10.1186/s12863-017-0514-6. BMC Genet. 2017. PMID: 28525980 Free PMC article.
References
-
- Mona S, Crestanello B, Bankhead-Dronnet S, Pecchioli E, Ingrosso S, D'Amelio S, Rossi L, Meneguz PG, Bertorelle G. Disentangling the effects of recombination, selection, and demography on the genetic variation at a major histocompatibility complex class II gene in the alpine chamois. Mol Ecol. 2008;13:13. - PubMed
-
- Hartl DL, Clark AG. Principles of population genetics. Sunderland, Massachusetts: Sinauer Associates, Inc; 1989.
-
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

