Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis
- PMID: 20682317
- PMCID: PMC2995815
- DOI: 10.1053/j.gastro.2010.07.050
Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis
Abstract
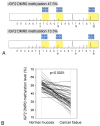
Background & aims: The insulin-like growth factor 2 (IGF2) gene is normally imprinted. Constitutive loss of imprinting (LOI) of IGF2 has been associated with increased risks of colon cancer and adenoma, indicating its role in carcinogenesis. The conventional LOI assay relies on a germline polymorphism to distinguish between 2 allelic expression patterns but results in many uninformative cases. IGF2 LOI correlates with hypomethylation at the differentially methylated region (DMR)-0. An assay for methylation of the DMR0 could overcome the limitations of the conventional IGF2 LOI assay.
Methods: We measured methylation at the IGF2 DMR0 using a bisulfite-pyrosequencing assay with 1178 paraffin-embedded colorectal cancer tissue samples from 2 prospective cohort studies. A Cox proportional hazard model was used to calculate mortality hazard ratio (HR); calculations were adjusted for microsatellite instability; the CpG island methylator phenotype; LINE-1 methylation; and KRAS, BRAF, and PIK3CA mutations.
Results: Methylation at the IGF2 DMR0 was successfully measured in 1105 (94%) of 1178 samples. Colorectal tumors had significantly less methylation at the DMR0 compared with matched, normal colonic mucosa (P < .0001; N = 51). Among 1033 patients eligible for survival analysis, hypomethylation of the IGF2 DMR0 was significantly associated with higher overall mortality (log-rank P = .0006; univariate HR, 1.41; 95% confidence interval, 1.16-1.71; P = .0006; multivariate HR, 1.33; 95% confidence interval, 1.08-1.63; P = .0066).
Conclusions: A bisulfite-pyrosequencing assay to measure methylation of the IGF2 DMR0 is robust and applicable to paraffin-embedded tissue. IGF2 DMR0 hypomethylation in colorectal tumor samples is associated with shorter survival time, so it might be developed as a prognostic biomarker.
Copyright © 2010 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–28. - PubMed
-
- Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–8. - PubMed
-
- Harper J, Burns JL, Foulstone EJ, et al. Soluble IGF2 receptor rescues Apc(Min/+) intestinal adenoma progression induced by Igf2 loss of imprinting. Cancer Res. 2006;66:1940–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

