Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype
- PMID: 20682644
- PMCID: PMC2955865
- DOI: 10.1158/1535-7163.MCT-10-0108
Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype
Abstract
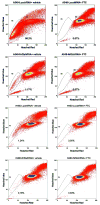
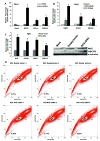
ATP-binding cassette, subfamily G, member 2 (ABCG2) is expressed in both normal and cancer cells and plays a crucial role in side population (SP) formation and efflux of xenobiotics and drugs. Nrf2, a redox-sensing transcription factor, on constitutive activation in non-small-cell lung cancer cells upregulates a wide spectrum of genes involved in redox balance, glutathione metabolism, and drug detoxification, which contribute to chemoresistance and tumorigenicity. This study examined the mechanism underlying Nrf2-dependent expression of ABCG2 and its role in the multidrug resistance phenotype. In silico analysis of the 5'-promoter flanking region of ABCG2 identified an antioxidant response element (ARE) at -431 to -420 bp. A detailed promoter analysis using luciferase reporter assays showed that ARE at -431 to -420 bp is critical for the Nrf2-mediated expression in lung cancer cells. Electrophoretic mobility shift assays and chromatin immunoprecipitation assays revealed that Nrf2 interacts with the ABCG2 ARE element at -431 to -420 bp in vitro and in vivo. Disruption of Nrf2 expression in lung and prostate cancer cells, by short hairpin RNA, attenuated the expression of ABCG2 transcript and protein, and dramatically reduced the SP fraction in Nrf2-depleted cancer cells. Moreover, depleted levels of ABCG2 in these Nrf2 knockdown cells sensitized them to mitoxantrone and topotecan, two chemotherapy drugs detoxified mainly by ABCG2. As expected, overexpression of Nrf2 cDNA in lung epithelial cells led to an increase in ABCG2 expression and a 2-fold higher SP fraction. Thus, Nrf2-mediated regulation of ABCG2 expression maintains the SP fraction and confers chemoresistance.
(c) 2010 AACR.
Conflict of interest statement
Figures




Similar articles
-
Elevated Expression of Nrf-2 and ABCG2 Involved in Multi-drug Resistance of Lung Cancer SP Cells.Drug Res (Stuttg). 2015 Oct;65(10):526-31. doi: 10.1055/s-0034-1390458. Epub 2014 Nov 4. Drug Res (Stuttg). 2015. PMID: 25368906
-
Transcription factors Sp1 and Sp3 regulate expression of human ABCG2 gene and chemoresistance phenotype.Mol Cells. 2013 Oct;36(4):368-75. doi: 10.1007/s10059-013-0191-x. Epub 2013 Aug 29. Mol Cells. 2013. PMID: 23996530 Free PMC article.
-
Transcriptional modulation of BCRP gene to reverse multidrug resistance by toremifene in breast adenocarcinoma cells.Breast Cancer Res Treat. 2010 Oct;123(3):679-89. doi: 10.1007/s10549-009-0660-2. Epub 2009 Dec 6. Breast Cancer Res Treat. 2010. PMID: 19967559
-
Multidrug resistance in cancer chemotherapy and xenobiotic protection mediated by the half ATP-binding cassette transporter ABCG2.Curr Med Chem Anticancer Agents. 2004 Jan;4(1):31-42. doi: 10.2174/1568011043482205. Curr Med Chem Anticancer Agents. 2004. PMID: 14754410 Review.
-
Role of Nrf2 in cancer photodynamic therapy: regulation of human ABC transporter ABCG2.J Pharm Sci. 2013 Sep;102(9):3058-69. doi: 10.1002/jps.23563. Epub 2013 May 6. J Pharm Sci. 2013. PMID: 23650051 Review.
Cited by
-
NRF2 Knockdown Resensitizes 5-Fluorouracil-Resistant Pancreatic Cancer Cells by Suppressing HO-1 and ABCG2 Expression.Int J Mol Sci. 2020 Jun 30;21(13):4646. doi: 10.3390/ijms21134646. Int J Mol Sci. 2020. PMID: 32629871 Free PMC article.
-
Bleomycin Induces Drug Efflux in Lungs. A Pitfall for Pharmacological Studies of Pulmonary Fibrosis.Am J Respir Cell Mol Biol. 2020 Feb;62(2):178-190. doi: 10.1165/rcmb.2018-0147OC. Am J Respir Cell Mol Biol. 2020. PMID: 31419911 Free PMC article.
-
Cytoprotection "gone astray": Nrf2 and its role in cancer.Onco Targets Ther. 2014 Aug 26;7:1497-518. doi: 10.2147/OTT.S36624. eCollection 2014. Onco Targets Ther. 2014. PMID: 25210464 Free PMC article. Review.
-
Montelukast ameliorated pemetrexed-induced cytotoxicity in hepatocytes by mitigating endoplasmic reticulum (ER) stress and nucleotide oligomerization domain-like receptor protein 3 (NLRP3) activation.Bioengineered. 2022 Mar;13(3):7894-7903. doi: 10.1080/21655979.2022.2051689. Bioengineered. 2022. PMID: 35291928 Free PMC article.
-
Breast cancer resistance protein (BCRP/ABCG2): its role in multidrug resistance and regulation of its gene expression.Chin J Cancer. 2012 Feb;31(2):73-99. doi: 10.5732/cjc.011.10320. Epub 2011 Nov 18. Chin J Cancer. 2012. PMID: 22098950 Free PMC article. Review.
References
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Wulf GG, Wang RY, Kuehnle I, et al. A leukemic stem cell with intrinsic drug efflux capacity in acute myeloid leukemia. Blood. 2001;98:1166–73. - PubMed
-
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

