Characterization of O-phosphohydroxyproline in rat {alpha}-crystallin A
- PMID: 20682783
- PMCID: PMC2951222
- DOI: 10.1074/jbc.M109.035428
Characterization of O-phosphohydroxyproline in rat {alpha}-crystallin A
Abstract
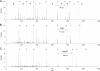
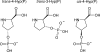
Post-translational modifications have major importance for the structure and function of a multiplicity of proteins. Phosphorylation is a widespread phenomenon among eukaryotic proteins. Whereas O-phosphorylation on the side chains of serine, threonine, and tyrosine in proteins is well known and has been studied extensively, to our knowledge the endogenous phosphorylation of hydroxyproline has not previously been reported. In the present work, we provide evidence for the first time that O-phosphohydroxyproline (Hyp(P)) is a proteinogenic amino acid. To detect Hyp(P) in proteins we generated a Hyp(P)-specific polyclonal antibody. We could identify Hyp(P) in various proteins by Western blot analysis, and we characterized the sequence position of Hyp(P) in the protein α-crystallin A by electrospray ionization-tandem mass spectrometry. Our experiments clearly demonstrate hydroxylation and subsequent phosphorylation of a proline residue in α-crystallin A in the eye as well as in heart tissue of rat.
Figures


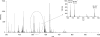

Similar articles
-
Phosphorylation site identification via ion trap tandem mass spectrometry of whole protein and peptide ions: bovine alpha-crystallin A chain.Anal Chem. 2003 Dec 1;75(23):6509-16. doi: 10.1021/ac034410s. Anal Chem. 2003. PMID: 14640721
-
Study of posttranslational modifications in lenticular alphaA-Crystallin of mice using proteomic analysis techniques.Biochim Biophys Acta. 2006 Dec;1764(12):1948-62. doi: 10.1016/j.bbapap.2006.10.004. Epub 2006 Oct 14. Biochim Biophys Acta. 2006. PMID: 17157567
-
Identification of phosphorylation and acetylation sites in alphaA-crystallin of the eye lens ( mus musculus) after two-dimensional gel electrophoresis.Anal Bioanal Chem. 2003 Aug;376(7):966-72. doi: 10.1007/s00216-003-1983-1. Epub 2003 Jul 10. Anal Bioanal Chem. 2003. PMID: 12856102
-
The implications of (2S,4S)-hydroxyproline 4-O-glycosylation for prolyl amide isomerization.Chemistry. 2009 Oct 12;15(40):10649-57. doi: 10.1002/chem.200900844. Chemistry. 2009. PMID: 19739208
-
Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth.Amino Acids. 2018 Jan;50(1):29-38. doi: 10.1007/s00726-017-2490-6. Epub 2017 Sep 20. Amino Acids. 2018. PMID: 28929384 Review.
Cited by
-
Proline editing: a general and practical approach to the synthesis of functionally and structurally diverse peptides. Analysis of steric versus stereoelectronic effects of 4-substituted prolines on conformation within peptides.J Am Chem Soc. 2013 Mar 20;135(11):4333-63. doi: 10.1021/ja3109664. Epub 2013 Mar 11. J Am Chem Soc. 2013. PMID: 23402492 Free PMC article.
-
Large-Scale Differentiation and Site Specific Discrimination of Hydroxyproline Isomers by Electron Transfer/Higher-Energy Collision Dissociation (EThcD) Mass Spectrometry.Anal Chem. 2018 May 1;90(9):5857-5864. doi: 10.1021/acs.analchem.8b00413. Epub 2018 Apr 20. Anal Chem. 2018. PMID: 29624053 Free PMC article.
References
-
- Hubbard M. J., Cohen P. (1993) Trends Biochem. Sci. 18, 172–177 - PubMed
-
- Kivirikko K. I., Pihlajaniemi T. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 325–398 - PubMed
-
- Francis S. H., Corbin J. D. (1994) Annu. Rev. Physiol. 56, 237–272 - PubMed
-
- Fantl W. J., Johnson D. E., Williams L. T. (1993) Annu. Rev. Biochem. 62, 453–481 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

