Root secretion of defense-related proteins is development-dependent and correlated with flowering time
- PMID: 20682788
- PMCID: PMC2945560
- DOI: 10.1074/jbc.M110.119040
Root secretion of defense-related proteins is development-dependent and correlated with flowering time
Abstract
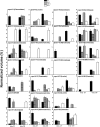
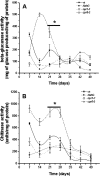
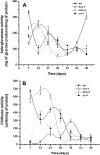
Proteins found in the root exudates are thought to play a role in the interactions between plants and soil organisms. To gain a better understanding of protein secretion by roots, we conducted a systematic proteomic analysis of the root exudates of Arabidopsis thaliana at different plant developmental stages. In total, we identified 111 proteins secreted by roots, the majority of which were exuded constitutively during all stages of development. However, defense-related proteins such as chitinases, glucanases, myrosinases, and others showed enhanced secretion during flowering. Defense-impaired mutants npr1-1 and NahG showed lower levels of secretion of defense proteins at flowering compared with the wild type. The flowering-defective mutants fca-1, stm-4, and co-1 showed almost undetectable levels of defense proteins in their root exudates at similar time points. In contrast, root secretions of defense-enhanced cpr5-2 mutants showed higher levels of defense proteins. The proteomics data were positively correlated with enzymatic activity assays for defense proteins and with in silico gene expression analysis of genes specifically expressed in roots of Arabidopsis. In conclusion, our results show a clear correlation between defense-related proteins secreted by roots and flowering time.
Figures



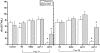
Similar articles
-
Root-secreted (-)-loliolide modulates both belowground defense and aboveground flowering in Arabidopsis and tobacco.J Exp Bot. 2023 Feb 5;74(3):964-975. doi: 10.1093/jxb/erac439. J Exp Bot. 2023. PMID: 36342376
-
Expression of a plant cell wall invertase in roots of Arabidopsis leads to early flowering and an increase in whole plant biomass.Plant Biol (Stuttg). 2005 Sep;7(5):469-75. doi: 10.1055/s-2005-865894. Plant Biol (Stuttg). 2005. PMID: 16163611
-
Overexpression of a phosphate transporter gene ZmPt9 from maize influences growth of transgenic Arabidopsis thaliana.Biochem Biophys Res Commun. 2021 Jun 18;558:196-201. doi: 10.1016/j.bbrc.2020.09.039. Epub 2020 Sep 19. Biochem Biophys Res Commun. 2021. PMID: 32962860
-
The control of flowering time by environmental factors.Plant J. 2017 May;90(4):708-719. doi: 10.1111/tpj.13461. Epub 2017 Feb 11. Plant J. 2017. PMID: 27995671 Review.
-
Tackling Plant Phosphate Starvation by the Roots.Dev Cell. 2019 Mar 11;48(5):599-615. doi: 10.1016/j.devcel.2019.01.002. Dev Cell. 2019. PMID: 30861374 Review.
Cited by
-
Metabolic niches in the rhizosphere microbiome: dependence on soil horizons, root traits and climate variables in forest ecosystems.Front Plant Sci. 2024 Apr 5;15:1344205. doi: 10.3389/fpls.2024.1344205. eCollection 2024. Front Plant Sci. 2024. PMID: 38645395 Free PMC article. Review.
-
Rhizosphere microbiome assemblage is affected by plant development.ISME J. 2014 Apr;8(4):790-803. doi: 10.1038/ismej.2013.196. Epub 2013 Nov 7. ISME J. 2014. PMID: 24196324 Free PMC article.
-
Engineering multifunctional rhizosphere probiotics using consortia of Bacillus amyloliquefaciens transposon insertion mutants.Elife. 2023 Sep 14;12:e90726. doi: 10.7554/eLife.90726. Elife. 2023. PMID: 37706503 Free PMC article.
-
Putative LysM Effectors Contribute to Fungal Lifestyle.Int J Mol Sci. 2021 Mar 19;22(6):3147. doi: 10.3390/ijms22063147. Int J Mol Sci. 2021. PMID: 33808705 Free PMC article.
-
Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions.PLoS One. 2013;8(2):e55731. doi: 10.1371/journal.pone.0055731. Epub 2013 Feb 1. PLoS One. 2013. PMID: 23383346 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

