The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth
- PMID: 20682797
- PMCID: PMC4261152
- DOI: 10.1158/0008-5472.CAN-10-0413
The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth
Abstract
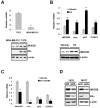
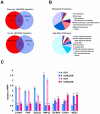
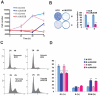
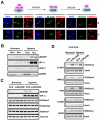
Estrogen receptor alpha (ERalpha) plays an important role in breast cancer. Upregulation of HIF-1alpha in ER(alpha)-positive cancers suggests that HIF-1alpha may cooperate with ERalpha to promote breast cancer progression and consequently affect breast cancer treatment. Here, we show the histone demethylase JMJD2B is regulated by both ERalpha and HIF-1alpha, drives breast cancer cell proliferation in normoxia and hypoxia, and epigenetically regulates the expression of cell cycle genes such as CCND1, CCNA1, and WEE1. We also show that JMJD2B and the hypoxia marker CA9 together stratify a subclass of breast cancer patients and predict a worse outcome of these breast cancers. Our findings provide a biological rationale to support the therapeutic targeting of histone demethylases in breast cancer patients.
Figures

Similar articles
-
HIF-1α-induced histone demethylase JMJD2B contributes to the malignant phenotype of colorectal cancer cells via an epigenetic mechanism.Carcinogenesis. 2012 Sep;33(9):1664-73. doi: 10.1093/carcin/bgs217. Epub 2012 Jun 27. Carcinogenesis. 2012. PMID: 22745382
-
miR-491-5p functions as a tumor suppressor by targeting JMJD2B in ERα-positive breast cancer.FEBS Lett. 2015 Mar 24;589(7):812-21. doi: 10.1016/j.febslet.2015.02.014. Epub 2015 Feb 25. FEBS Lett. 2015. PMID: 25725194
-
Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development.PLoS One. 2011 Mar 18;6(3):e17830. doi: 10.1371/journal.pone.0017830. PLoS One. 2011. PMID: 21445275 Free PMC article.
-
Hypoxia and Hormone-Mediated Pathways Converge at the Histone Demethylase KDM4B in Cancer.Int J Mol Sci. 2018 Jan 13;19(1):240. doi: 10.3390/ijms19010240. Int J Mol Sci. 2018. PMID: 29342868 Free PMC article. Review.
-
Insights into The Function and Regulation of Jumonji C Lysine Demethylases as Hypoxic Responsive Enzymes.Curr Protein Pept Sci. 2020;21(7):642-654. doi: 10.2174/1389203721666191231104225. Curr Protein Pept Sci. 2020. PMID: 31889485 Review.
Cited by
-
Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation.J Biol Chem. 2013 Jul 19;288(29):21376-21388. doi: 10.1074/jbc.M113.491514. Epub 2013 Jun 6. J Biol Chem. 2013. PMID: 23744078 Free PMC article.
-
Chromatin as an oxygen sensor and active player in the hypoxia response.Cell Signal. 2012 Jan;24(1):35-43. doi: 10.1016/j.cellsig.2011.08.019. Epub 2011 Sep 8. Cell Signal. 2012. PMID: 21924352 Free PMC article. Review.
-
Hypoxia-induced epigenetic regulation of breast cancer progression and the tumour microenvironment.Front Cell Dev Biol. 2024 Aug 30;12:1421629. doi: 10.3389/fcell.2024.1421629. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39282472 Free PMC article. Review.
-
KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells.Cancer Res. 2013 May 15;73(10):2936-42. doi: 10.1158/0008-5472.CAN-12-4300. Epub 2013 May 3. Cancer Res. 2013. PMID: 23644528 Free PMC article. Review.
-
Histone demethylases and their roles in cancer epigenetics.J Med Oncol Ther. 2016;1(2):34-40. J Med Oncol Ther. 2016. PMID: 28149961 Free PMC article.
References
-
- Enmark E, Gustafsson JA. Oestrogen receptors - an overview. J Intern Med. 1999;246:133–8. - PubMed
-
- McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–4. - PubMed
-
- Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006;20:1707–14. - PubMed
-
- Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–78. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

