Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation
- PMID: 20682837
- PMCID: PMC2947163
- DOI: 10.1105/tpc.110.076075
Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation
Abstract
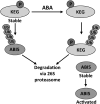
The Arabidopsis thaliana RING-type E3 ligase KEEP ON GOING (KEG) is a negative regulator of abscisic acid (ABA) signaling. Seedlings homozygous for T-DNA insertions in KEG accumulate high levels of the ABA-responsive transcription factor ABSCISIC ACID-INSENSITIVE5 (ABI5). Here, we demonstrate that KEG E3 ligase activity is required for the regulation of ABI5 abundance. KEG ubiquitinates ABI5 in vitro, and a functional KEG RING domain is required to restore the levels of ABI5 in keg-1 to that of the wild type. Overexpression of KEG leads to ABA insensitivity, which correlates with KEG protein levels. In the presence of ABA, ABI5 levels increase drastically via a decrease in ubiquitin-meditated proteasomal degradation. Our results indicate that ABA promotes ABI5 accumulation by inducing the ubiquitination and proteasomal degradation of KEG. A functional RING domain is required for the ABA-induced degradation of KEG, suggesting that the loss is due to self-ubiquitination. Mutations within KEG's kinase domain or treatments with kinase inhibitors prohibit the ABA-induced ubiquitination and degradation of KEG, indicating that phosphorylation, possibly self-phosphorylation, is involved in the ABA regulation of KEG protein levels. We discuss a model for how ABA may negatively regulate KEG protein abundance, leading to accumulation of ABI5 and ABA-dependent cellular responses.
Figures






Similar articles
-
KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling.Plant Cell. 2006 Dec;18(12):3415-28. doi: 10.1105/tpc.106.046532. Epub 2006 Dec 28. Plant Cell. 2006. PMID: 17194765 Free PMC article.
-
Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING.J Biol Chem. 2013 Jul 12;288(28):20267-79. doi: 10.1074/jbc.M113.465369. Epub 2013 May 29. J Biol Chem. 2013. PMID: 23720747 Free PMC article.
-
Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system.J Exp Bot. 2013 Jul;64(10):2779-91. doi: 10.1093/jxb/ert123. Epub 2013 May 8. J Exp Bot. 2013. PMID: 23658427 Free PMC article.
-
Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants.Cells. 2021 Aug 5;10(8):1996. doi: 10.3390/cells10081996. Cells. 2021. PMID: 34440762 Free PMC article. Review.
-
Structural Dynamics, Evolutionary Significance, and Functions of Really Interesting New Gene Proteins in Ubiquitination and Plant Stress: A Review.DNA Cell Biol. 2025 May;44(5):214-228. doi: 10.1089/dna.2025.0002. Epub 2025 Apr 10. DNA Cell Biol. 2025. PMID: 40208634 Review.
Cited by
-
Integration of ABA, GA, and light signaling in seed germination through the regulation of ABI5.Front Plant Sci. 2022 Aug 24;13:1000803. doi: 10.3389/fpls.2022.1000803. eCollection 2022. Front Plant Sci. 2022. PMID: 36092418 Free PMC article. Review.
-
Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis.Plant Cell. 2013 Feb;25(2):625-36. doi: 10.1105/tpc.112.106880. Epub 2013 Feb 19. Plant Cell. 2013. PMID: 23424245 Free PMC article.
-
ABA signaling prevents phosphodegradation of the SR45 splicing factor to alleviate inhibition of early seedling development in Arabidopsis.Plant Commun. 2023 Mar 13;4(2):100495. doi: 10.1016/j.xplc.2022.100495. Epub 2022 Nov 23. Plant Commun. 2023. PMID: 36419364 Free PMC article.
-
CRISPR/Cas9 Gene Editing of NtAITRs, a Family of Transcription Repressor Genes, Leads to Enhanced Drought Tolerance in Tobacco.Int J Mol Sci. 2022 Dec 3;23(23):15268. doi: 10.3390/ijms232315268. Int J Mol Sci. 2022. PMID: 36499605 Free PMC article.
-
Wheat F-box Protein TaFBA1 Positively Regulates Plant Drought Tolerance but Negatively Regulates Stomatal Closure.Front Plant Sci. 2019 Oct 10;10:1242. doi: 10.3389/fpls.2019.01242. eCollection 2019. Front Plant Sci. 2019. PMID: 31649704 Free PMC article.
References
-
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Chico J.M., Chini A., Fonseca S., Solano R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11: 486–494 - PubMed
-
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

