Active DNA demethylation: many roads lead to Rome
- PMID: 20683471
- PMCID: PMC3711520
- DOI: 10.1038/nrm2950
Active DNA demethylation: many roads lead to Rome
Erratum in
- Nat Rev Mol Cell Biol. 2010 Oct;11(10):750
Abstract
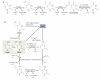
DNA methylation is one of the best-characterized epigenetic modifications and has been implicated in numerous biological processes, including transposable element silencing, genomic imprinting and X chromosome inactivation. Compared with other epigenetic modifications, DNA methylation is thought to be relatively stable. Despite its role in long-term silencing, DNA methylation is more dynamic than originally thought as active DNA demethylation has been observed during specific stages of development. In the past decade, many enzymes have been proposed to carry out active DNA demethylation and growing evidence suggests that, depending on the context, this process may be achieved by multiple mechanisms. Insight into how DNA methylation is dynamically regulated will broaden our understanding of epigenetic regulation and have great implications in somatic cell reprogramming and regenerative medicine.
Figures
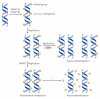
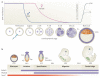
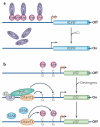
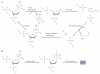
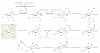
References
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 2003;33:245–254. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Rev. Genet. 2007;8:286–298. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Rev. Cancer. 2004;4:143–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

