Analysis of plant diversity with retrotransposon-based molecular markers
- PMID: 20683483
- PMCID: PMC3183911
- DOI: 10.1038/hdy.2010.93
Analysis of plant diversity with retrotransposon-based molecular markers
Abstract
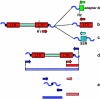
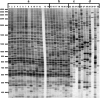
Retrotransposons are both major generators of genetic diversity and tools for detecting the genomic changes associated with their activity because they create large and stable insertions in the genome. After the demonstration that retrotransposons are ubiquitous, active and abundant in plant genomes, various marker systems were developed to exploit polymorphisms in retrotransposon insertion patterns. These have found applications ranging from the mapping of genes responsible for particular traits and the management of backcrossing programs to analysis of population structure and diversity of wild species. This review provides an insight into the spectrum of retrotransposon-based marker systems developed for plant species and evaluates the contributions of retrotransposon markers to the analysis of population diversity in plants.
Figures


Similar articles
-
Use of retrotransposon-derived genetic markers to analyse genomic variability in plants.Funct Plant Biol. 2018 Jan;46(1):15-29. doi: 10.1071/FP18098. Funct Plant Biol. 2018. PMID: 30939255 Review.
-
Diversity of the Ty-1 copia retrotransposon Tos17 in rice (Oryza sativa L.) and the AA genome of the Oryza genus.Mol Genet Genomics. 2009 Dec;282(6):633-52. doi: 10.1007/s00438-009-0493-z. Epub 2009 Oct 25. Mol Genet Genomics. 2009. PMID: 19856189
-
Genetic bottlenecks in Turkish okra germplasm and utility of iPBS retrotransposon markers for genetic diversity assessment.Genet Mol Res. 2015 Sep 8;14(3):10588-602. doi: 10.4238/2015.September.8.20. Genet Mol Res. 2015. PMID: 26400290
-
Polymorphisms and evolutionary history of retrotransposon insertions in rice promoters.Genome. 2011 Aug;54(8):629-38. doi: 10.1139/g11-030. Epub 2011 Aug 8. Genome. 2011. PMID: 21823826
-
Parasitism and the retrotransposon life cycle in plants: a hitchhiker's guide to the genome.Heredity (Edinb). 2006 Dec;97(6):381-8. doi: 10.1038/sj.hdy.6800903. Epub 2006 Sep 20. Heredity (Edinb). 2006. PMID: 16985508 Review.
Cited by
-
Differential dynamics of transposable elements during long-term diploidization of Nicotiana section Repandae (Solanaceae) allopolyploid genomes.PLoS One. 2012;7(11):e50352. doi: 10.1371/journal.pone.0050352. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185607 Free PMC article.
-
Obtaining retrotransposon sequences, analysis of their genomic distribution and use of retrotransposon-derived genetic markers in lentil (Lens culinaris Medik.).PLoS One. 2017 Apr 27;12(4):e0176728. doi: 10.1371/journal.pone.0176728. eCollection 2017. PLoS One. 2017. PMID: 28448614 Free PMC article.
-
Using two retrotransposon-based marker systems (SRAP and REMAP) for genetic diversity analysis of Moroccan Argan tree.Mol Biol Res Commun. 2020 Sep;9(3):93-103. doi: 10.22099/mbrc.2020.36390.1478. Mol Biol Res Commun. 2020. PMID: 33313328 Free PMC article.
-
Characterization and potential evolutionary impact of transposable elements in the genome of Cochliobolus heterostrophus.BMC Genomics. 2014 Jun 28;15(1):536. doi: 10.1186/1471-2164-15-536. BMC Genomics. 2014. PMID: 24973942 Free PMC article.
-
Comparative Study of Pine Reference Genomes Reveals Transposable Element Interconnected Gene Networks.Genes (Basel). 2020 Oct 16;11(10):1216. doi: 10.3390/genes11101216. Genes (Basel). 2020. PMID: 33081418 Free PMC article.
References
-
- Antonius-Klemola K, Kalendar R, Schulman AH. TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor Appl Genet. 2006;112:999–1008. - PubMed
-
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Baumel A, Ainouche M, Kalendar R, Schulman AH. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae) Mol Biol Evol. 2002;19:1218–1227. - PubMed
-
- Bernet GP, Asins MJ. Identification and genomic distribution of gypsy like retrotransposons in Citrus and Poncirus. Theor Appl Genet. 2004;108:121–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

