Prediction of the three-dimensional structure for the rat urotensin II receptor, and comparison of the antagonist binding sites and binding selectivity between human and rat receptors from atomistic simulations
- PMID: 20683923
- PMCID: PMC3517062
- DOI: 10.1002/cmdc.201000175
Prediction of the three-dimensional structure for the rat urotensin II receptor, and comparison of the antagonist binding sites and binding selectivity between human and rat receptors from atomistic simulations
Abstract
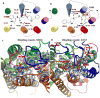
Urotensin-II (U-II) has been shown to be the most potent mammalian vasoconstrictor known. Thus, a U-II antagonist might be of therapeutic value in a number of cardiovascular disorders. However, interspecies variability of several nonpeptidic ligands complicates the interpretation of in vivo studies of such antagonists in preclinical animal disease models. ACT058362 is a selective antagonist for the human U-II receptor (hUT2R) with a reported K(d) value of approximately 4 nM in a molecular binding assay, but it is reported to bind weakly to rat UT2R (rUT2R), with a K(d) value of approximately 1 500 nM. In contrast, the arylsulphonamide SB706375 is a selective antagonist against both hUT2R (K(d)= approximately 9 nM) and rUT2R (K(d)= approximately 21 nM). To understand the species selectivity of the UT2R, we investigated the binding site of ACT058362 and SB706375 in both hUT2R and rUT2R to explain the dramatically lower (approximately 400-fold) affinity of ACT058362 for rUT2R and the similar affinity (approximately 10 nM) of SB706375 for both UT2Rs. These studies used MembStruk and MSCDock to predict the UT2R structure and the binding site of ACT058362 and SB706375. Based on binding energies, we found two binding modes each with D130(3.32) as the crucial anchoring point (Ballesteros-Weinstein numbering given in superscript). We predict that ACT058362 (an aryl-amine-aryl or ANA ligand) binds in the transmembrane (TM) 3456 region, while SB706375 (an aryl-aryl-amine or AAN ligand) binds in the TM 1237 region. These predicted sites explain the known differences in binding of the ANA ligand to rat and human receptors, while explaining the similar binding of the AAN compound to rat and human receptors. Moreover the predictions explain currently available structure-activity relationship (SAR) data. To further validate the predicted binding sites of these ligands in hUT2R and rUT2R, we propose several mutations that would help define the structural origins of differential responses between UT2R of different species, potentially indicating novel UT2R antagonists with cross-species high affinity.
Figures





Similar articles
-
Predicted ligands for the human urotensin-II G protein-coupled receptor with some experimental validation.ChemMedChem. 2014 Aug;9(8):1732-43. doi: 10.1002/cmdc.201402087. Epub 2014 Jul 2. ChemMedChem. 2014. PMID: 24989481
-
Three-dimensional model of the human urotensin-II receptor: docking of human urotensin-II and nonpeptide antagonists in the binding site and comparison with an antagonist pharmacophore model.Proteins. 2008 Oct;73(1):173-84. doi: 10.1002/prot.22050. Proteins. 2008. PMID: 18409194
-
Pharmacology of the urotensin-II receptor antagonist palosuran (ACT-058362; 1-[2-(4-benzyl-4-hydroxy-piperidin-1-yl)-ethyl]-3-(2-methyl-quinolin-4-yl)-urea sulfate salt): first demonstration of a pathophysiological role of the urotensin System.J Pharmacol Exp Ther. 2004 Oct;311(1):204-12. doi: 10.1124/jpet.104.068320. Epub 2004 May 14. J Pharmacol Exp Ther. 2004. PMID: 15146030
-
Nonpeptide Urotensin-II receptor agonists and antagonists: review and structure-activity relationships.Peptides. 2008 May;29(5):680-90. doi: 10.1016/j.peptides.2007.09.019. Epub 2007 Oct 6. Peptides. 2008. PMID: 18022732 Review.
-
Relaxin-3, INSL5, and their receptors.Results Probl Cell Differ. 2008;46:213-37. doi: 10.1007/400_2007_055. Results Probl Cell Differ. 2008. PMID: 18236022 Review.
Cited by
-
Predicted structures of agonist and antagonist bound complexes of adenosine A3 receptor.Proteins. 2011 Jun;79(6):1878-97. doi: 10.1002/prot.23012. Epub 2011 Apr 12. Proteins. 2011. PMID: 21488099 Free PMC article.
-
Characterizing and predicting the functional and conformational diversity of seven-transmembrane proteins.Methods. 2011 Dec;55(4):405-14. doi: 10.1016/j.ymeth.2011.12.005. Epub 2011 Dec 17. Methods. 2011. PMID: 22197575 Free PMC article.
-
Predicted 3D structures of olfactory receptors with details of odorant binding to OR1G1.J Comput Aided Mol Des. 2014 Dec;28(12):1175-90. doi: 10.1007/s10822-014-9793-4. Epub 2014 Sep 16. J Comput Aided Mol Des. 2014. PMID: 25224127
-
Selectivity and activation of dopamine D3R from molecular dynamics.J Mol Model. 2012 Dec;18(12):5051-63. doi: 10.1007/s00894-012-1509-x. Epub 2012 Jul 3. J Mol Model. 2012. PMID: 22752545
-
Elucidating glycosaminoglycan-protein-protein interactions using carbohydrate microarray and computational approaches.Proc Natl Acad Sci U S A. 2011 Jun 14;108(24):9747-52. doi: 10.1073/pnas.1102962108. Epub 2011 May 31. Proc Natl Acad Sci U S A. 2011. PMID: 21628576 Free PMC article.
References
-
- Ames RS, Sarau HM, Chambers JK, Willette RN, Aiyar RV, Romanic AM, Louden CS, Foley JJ, Sauermelch CF, Coatney RW, Ao Z, Disa J, Holmes SD, Stadel JM, Martin JD, Liu WS, Glover GI, Wilson S, McNutty DE, Ellis CE, Eishourbagy NA, Shabon U, Trill JJ, Hay DVP, Ohlstein EH, Bergsma DJ, Douglas SA. Nature. 1999;401:282–286. - PubMed
-
- Douglas SA. Curr Opin Pharmacol. 2003;3:159–167. - PubMed
-
- Carotenuto A, Grieco P, Campiglia P, Novellino E, Rovero P. J Med Chem. 2004;47:1652–1661. - PubMed
-
- Clozel M, Binkert C, Birker-Robaczewska M, Boukhadra C, Ding S-S, Fischli W, Hess P, Mathys B, Morrison K, Müller C, Müller C, Nayler O, Qiu C, Rey M, Scherz MW, Velker J, Weller T, Zi J-F, Ziltenerm P. J Pharmacol Exp Ther. 2004;311:204–212. - PubMed
-
- Flohr S, Kurtz M, Kostenis E, Brkovich A, Fournier A, Klabunde T. J Med Chem. 2002;45:1799–1805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

