Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid
- PMID: 20684044
- PMCID: PMC4793595
- DOI: 10.1016/j.jmb.2010.05.058
Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid
Abstract
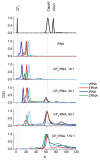
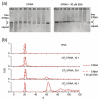
Single-stranded RNA viruses package their genomes into capsids enclosing fixed volumes. We assayed the ability of bacteriophage MS2 coat protein to package large, defined fragments of its genomic, single-stranded RNA. We show that the efficiency of packaging into a T=3 capsid in vitro is inversely proportional to RNA length, implying that there is a free-energy barrier to be overcome during assembly. All the RNAs examined have greater solution persistence lengths than the internal diameter of the capsid into which they become packaged, suggesting that protein-mediated RNA compaction must occur during assembly. Binding ethidium bromide to one of these RNA fragments, which would be expected to reduce its flexibility, severely inhibited packaging, consistent with this idea. Cryo-EM structures of the capsids assembled in these experiments with the sub-genomic RNAs show a layer of RNA density beneath the coat protein shell but lack density for the inner RNA shell seen in the wild-type virion. The inner layer is restored when full-length virion RNA is used in the assembly reaction, implying that it becomes ordered only when the capsid is filled, presumably because of the effects of steric and/or electrostatic repulsions. The cryo-EM results explain the length dependence of packaging. In addition, they show that for the sub-genomic fragments the strongest ordered RNA density occurs below the coat protein dimers forming the icosahedral 5-fold axes of the capsid. There is little such density beneath the proteins at the 2-fold axes, consistent with our model in which coat protein dimers binding to RNA stem-loops located at sites throughout the genome leads to switching of their preferred conformations, thus regulating the placement of the quasi-conformers needed to build the T=3 capsid. The data are consistent with mutual chaperoning of both RNA and coat protein conformations, partially explaining the ability of such viruses to assemble so rapidly and accurately.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures



References
-
- Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 2006;60:51–67. - PubMed
-
- Harrison SC, Olson AJ, Schutt CE, Winkler FK. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. - PubMed
-
- Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

