Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: modeling and experimental validation of a chimera peptide
- PMID: 20684611
- PMCID: PMC2917172
- DOI: 10.1021/jm1002974
Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: modeling and experimental validation of a chimera peptide
Abstract
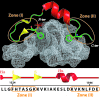
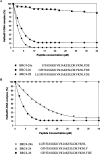
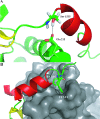
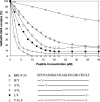
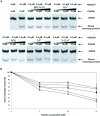
We have previously shown that a 28-amino acid peptide derived from the BRC4 motif of BRCA2 tumor suppressor inhibits selectively human RAD51 recombinase (HsRad51). With the aim of designing better inhibitors for cancer treatment, we combined an in silico docking approach with in vitro biochemical testing to construct a highly efficient chimera peptide from eight existing human BRC motifs. We built a molecular model of all BRC motifs complexed with HsRad51 based on the crystal structure of the BRC4 motif-HsRad51 complex, computed the interaction energy of each residue in each BRC motif, and selected the best amino acid residue at each binding position. This analysis enabled us to propose four amino acid substitutions in the BRC4 motif. Three of these increased the inhibitory effect in vitro, and this effect was found to be additive. We thus obtained a peptide that is about 10 times more efficient in inhibiting HsRad51-ssDNA complex formation than the original peptide.
Figures


References
-
- Friedberg E. C.; Walker G. C.; Siede W.; Wood R. D.; Schultz R. A.; Ellenberger T.. DNA Repair and Mutagenesis; ASM Press: Washington, DC, 2006.
-
- Christodoulopoulos G.; Malapetsa A.; Schipper H.; Golub E.; Radding C.; Panasci L. C. Chlorambucil induction of HsRad51 in B-cell chronic lymphocytic leukemia. Clin. Cancer Res. 1999, 5, 2178–2184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials
Miscellaneous

