Genome mapping and characterization of the Anopheles gambiae heterochromatin
- PMID: 20684766
- PMCID: PMC3091655
- DOI: 10.1186/1471-2164-11-459
Genome mapping and characterization of the Anopheles gambiae heterochromatin
Abstract
Background: Heterochromatin plays an important role in chromosome function and gene regulation. Despite the availability of polytene chromosomes and genome sequence, the heterochromatin of the major malaria vector Anopheles gambiae has not been mapped and characterized.
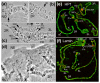
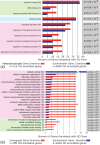
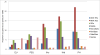
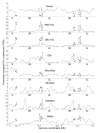
Results: To determine the extent of heterochromatin within the An. gambiae genome, genes were physically mapped to the euchromatin-heterochromatin transition zone of polytene chromosomes. The study found that a minimum of 232 genes reside in 16.6 Mb of mapped heterochromatin. Gene ontology analysis revealed that heterochromatin is enriched in genes with DNA-binding and regulatory activities. Immunostaining of the An. gambiae chromosomes with antibodies against Drosophila melanogaster heterochromatin protein 1 (HP1) and the nuclear envelope protein lamin Dm0 identified the major invariable sites of the proteins' localization in all regions of pericentric heterochromatin, diffuse intercalary heterochromatin, and euchromatic region 9C of the 2R arm, but not in the compact intercalary heterochromatin. To better understand the molecular differences among chromatin types, novel Bayesian statistical models were developed to analyze genome features. The study found that heterochromatin and euchromatin differ in gene density and the coverage of retroelements and segmental duplications. The pericentric heterochromatin had the highest coverage of retroelements and tandem repeats, while intercalary heterochromatin was enriched with segmental duplications. We also provide evidence that the diffuse intercalary heterochromatin has a higher coverage of DNA transposable elements, minisatellites, and satellites than does the compact intercalary heterochromatin. The investigation of 42-Mb assembly of unmapped genomic scaffolds showed that it has molecular characteristics similar to cytologically mapped heterochromatin.
Conclusions: Our results demonstrate that Anopheles polytene chromosomes and whole-genome shotgun assembly render the mapping and characterization of a significant part of heterochromatic scaffolds a possibility. These results reveal the strong association between characteristics of the genome features and morphological types of chromatin. Initial analysis of the An. gambiae heterochromatin provides a framework for its functional characterization and comparative genomic analyses with other organisms.
Figures


Similar articles
-
Genomic composition and evolution of Aedes aegypti chromosomes revealed by the analysis of physically mapped supercontigs.BMC Biol. 2014 Apr 14;12:27. doi: 10.1186/1741-7007-12-27. BMC Biol. 2014. PMID: 24731704 Free PMC article.
-
Distribution of T1, Q, Pegasus and mariner transposable elements on the polytene chromosomes of PEST, a standard strain of Anopheles gambiae.Chromosoma. 1996 Jun;104(8):585-95. doi: 10.1007/BF00352298. Chromosoma. 1996. PMID: 8662251
-
Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes.PLoS One. 2010 May 12;5(5):e10592. doi: 10.1371/journal.pone.0010592. PLoS One. 2010. PMID: 20485676 Free PMC article.
-
Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster.Chromosoma. 2008 Oct;117(5):411-8. doi: 10.1007/s00412-008-0163-7. Epub 2008 May 20. Chromosoma. 2008. PMID: 18491121 Review.
-
[Distorted heterochromatin replication in Drosophila melanogaster polytene chromosomes as a result of euchromatin-heterochromatin rearrangements].Genetika. 2007 Jan;43(1):18-26. Genetika. 2007. PMID: 17333934 Review. Russian.
Cited by
-
Structural Variation of the X Chromosome Heterochromatin in the Anopheles gambiae Complex.Genes (Basel). 2020 Mar 19;11(3):327. doi: 10.3390/genes11030327. Genes (Basel). 2020. PMID: 32204543 Free PMC article.
-
A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing.Genes (Basel). 2019 Jan 18;10(1):62. doi: 10.3390/genes10010062. Genes (Basel). 2019. PMID: 30669388 Free PMC article.
-
Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes.Genome Res. 2019 Aug;29(8):1250-1261. doi: 10.1101/gr.245795.118. Epub 2019 Jul 25. Genome Res. 2019. PMID: 31345938 Free PMC article.
-
Heterochromatin, histone modifications, and nuclear architecture in disease vectors.Curr Opin Insect Sci. 2015 Aug 1;10:110-117. doi: 10.1016/j.cois.2015.05.003. Curr Opin Insect Sci. 2015. PMID: 26097808 Free PMC article.
-
Anopheles mosquitoes reveal new principles of 3D genome organization in insects.Nat Commun. 2022 Apr 12;13(1):1960. doi: 10.1038/s41467-022-29599-5. Nat Commun. 2022. PMID: 35413948 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

