Systematic analysis of short internal indels and their impact on protein folding
- PMID: 20684774
- PMCID: PMC2924343
- DOI: 10.1186/1472-6807-10-24
Systematic analysis of short internal indels and their impact on protein folding
Abstract
Background: Protein sequence insertions/deletions (indels) can be introduced during evolution or through alternative splicing (AS). Alternative splicing is an important biological phenomenon and is considered as the major means of expanding structural and functional diversity in eukaryotes. Knowledge of the structural changes due to indels is critical to our understanding of the evolution of protein structure and function. In addition, it can help us probe the evolution of alternative splicing and the diversity of functional isoforms. However, little is known about the effects of indels, in particular the ones involving core secondary structures, on the folding of protein structures. The long term goal of our study is to accurately predict the protein AS isoform structures. As a first step towards this goal, we performed a systematic analysis on the structural changes caused by short internal indels through mining highly homologous proteins in Protein Data Bank (PDB).
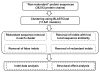
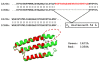
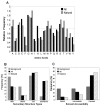
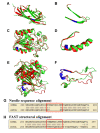
Results: We compiled a non-redundant dataset of short internal indels (2-40 amino acids) from highly homologous protein pairs and analyzed the sequence and structural features of the indels. We found that about one third of indel residues are in disordered state and majority of the residues are exposed to solvent, suggesting that these indels are generally located on the surface of proteins. Though naturally occurring indels are fewer than engineered ones in the dataset, there are no statistically significant differences in terms of amino acid frequencies and secondary structure types between the "Natural" indels and "All" indels in the dataset. Structural comparisons show that all the protein pairs with short internal indels in the dataset preserve the structural folds and about 85% of protein pairs have global RMSDs (root mean square deviations) of 2A or less, suggesting that protein structures tend to be conserved and can tolerate short insertions and deletions. A few pairs with high RMSDs are results of relative domain positions of the proteins, probably due to the intrinsically dynamic nature of the proteins.
Conclusions: The analysis demonstrated that protein structures have the "plasticity" to tolerate short indels. This study can provide valuable guides in modeling protein AS isoform structures and homologous proteins with indels through placing the indels at the right locations since the accuracy of sequence alignments dictate model qualities in homology modeling.
Figures




Similar articles
-
Indel PDB: a database of structural insertions and deletions derived from sequence alignments of closely related proteins.BMC Bioinformatics. 2008 Jun 25;9:293. doi: 10.1186/1471-2105-9-293. BMC Bioinformatics. 2008. PMID: 18578882 Free PMC article.
-
Protein expansion is primarily due to indels in intrinsically disordered regions.Mol Biol Evol. 2013 Dec;30(12):2645-53. doi: 10.1093/molbev/mst157. Epub 2013 Sep 12. Mol Biol Evol. 2013. PMID: 24037790
-
Long indels are disordered: a study of disorder and indels in homologous eukaryotic proteins.Biochim Biophys Acta. 2013 May;1834(5):890-7. doi: 10.1016/j.bbapap.2013.01.002. Epub 2013 Jan 17. Biochim Biophys Acta. 2013. PMID: 23333420
-
Insertions and Deletions (Indels): A Missing Piece of the Protein Engineering Jigsaw.Biochemistry. 2023 Jan 17;62(2):148-157. doi: 10.1021/acs.biochem.2c00188. Epub 2022 Jul 13. Biochemistry. 2023. PMID: 35830609 Review.
-
Insertions and deletions in protein evolution and engineering.Biotechnol Adv. 2022 Nov;60:108010. doi: 10.1016/j.biotechadv.2022.108010. Epub 2022 Jun 20. Biotechnol Adv. 2022. PMID: 35738511 Review.
Cited by
-
Fitness Effects of Single Amino Acid Insertions and Deletions in TEM-1 β-Lactamase.J Mol Biol. 2019 May 31;431(12):2320-2330. doi: 10.1016/j.jmb.2019.04.030. Epub 2019 Apr 26. J Mol Biol. 2019. PMID: 31034887 Free PMC article.
-
Elucidating the Structural Impacts of Protein InDels.Biomolecules. 2022 Oct 7;12(10):1435. doi: 10.3390/biom12101435. Biomolecules. 2022. PMID: 36291643 Free PMC article.
-
Computational analysis of pathogen-borne metallo β-lactamases reveals discriminating structural features between B1 types.BMC Res Notes. 2012 Feb 14;5:96. doi: 10.1186/1756-0500-5-96. BMC Res Notes. 2012. PMID: 22333139 Free PMC article.
-
Computational prediction of the tolerance to amino-acid deletion in green-fluorescent protein.PLoS One. 2017 Apr 3;12(4):e0164905. doi: 10.1371/journal.pone.0164905. eCollection 2017. PLoS One. 2017. PMID: 28369116 Free PMC article.
-
Local structural differences in homologous proteins: specificities in different SCOP classes.PLoS One. 2012;7(6):e38805. doi: 10.1371/journal.pone.0038805. Epub 2012 Jun 22. PLoS One. 2012. PMID: 22745680 Free PMC article.
References
-
- Pennisi E. Why do humans have so few genes? Science (New York, NY) 2005;309(5731):80.. - PubMed
-
- Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PI, Albrecht M, Hegyi H, Giorgetti A, Raimondo D, Lagarde J, Laskowski RA, Lopez G, Sadowski MI, Watson JD, Fariselli P, Rossi I, Nagy A, Kai W, Storling Z, Orsini M, Assenov Y, Blankenburg H, Huthmacher C, Ramirez F, Schlicker A, Denoeud F, Jones P, Kerrien S. The implications of alternative splicing in the ENCODE protein complement. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5495–5500. doi: 10.1073/pnas.0700800104. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

