Dynamic fluorescence depolarization: a powerful tool to explore protein folding on the ribosome
- PMID: 20685617
- PMCID: PMC2934862
- DOI: 10.1016/j.ymeth.2010.06.001
Dynamic fluorescence depolarization: a powerful tool to explore protein folding on the ribosome
Abstract
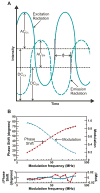
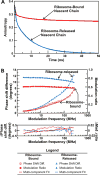
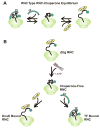
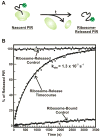
Protein folding is a fundamental biological process of great significance for cell function and life-related processes. Surprisingly, very little is presently known about how proteins fold in vivo. The influence of the cellular environment is of paramount importance, as molecular chaperones, the ribosome, and the crowded medium affect both folding pathways and potentially even equilibrium structures. Studying protein folding in physiologically relevant environments, however, poses a number of technical challenges due to slow tumbling rates, low concentrations and potentially non-homogenous populations. Early work in this area relied on biological assays based on antibody recognition, proteolysis, and activity studies. More recently, it has been possible to directly observe the structure and dynamics of nascent polypeptides at high resolution by spectroscopic and microscopic techniques. The fluorescence depolarization decay of nascent polypeptides labeled with a small extrinsic fluorophore is a particularly powerful tool to gain insights into the dynamics of newly synthesized proteins. The fluorophore label senses both its own local mobility and the motions of the macromolecule to which it is attached. Fluorescence anisotropy decays can be measured both in the time and frequency domains. The latter mode of data collection is extremely convenient to capture the nanosecond motions in ribosome-bound nascent proteins, indicative of the development of independent structure and folding on the ribosome. In this review, we discuss the theory of fluorescence depolarization and its exciting applications to the study of the dynamics of nascent proteins in the cellular environment.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures

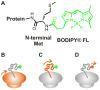




Similar articles
-
Confined dynamics of a ribosome-bound nascent globin: Cone angle analysis of fluorescence depolarization decays in the presence of two local motions.Protein Sci. 2009 Oct;18(10):2003-15. doi: 10.1002/pro.196. Protein Sci. 2009. PMID: 19569194 Free PMC article.
-
Fluorescence Anisotropy Decays and Microscale-Volume Viscometry Reveal the Compaction of Ribosome-Bound Nascent Proteins.J Phys Chem B. 2021 Jun 24;125(24):6543-6558. doi: 10.1021/acs.jpcb.1c04473. Epub 2021 Jun 10. J Phys Chem B. 2021. PMID: 34110829 Free PMC article.
-
Chain dynamics of nascent polypeptides emerging from the ribosome.ACS Chem Biol. 2008 Sep 19;3(9):555-66. doi: 10.1021/cb800059u. Epub 2008 Aug 22. ACS Chem Biol. 2008. PMID: 18717565 Free PMC article.
-
The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins.Nat Struct Mol Biol. 2009 Jun;16(6):589-97. doi: 10.1038/nsmb.1614. Nat Struct Mol Biol. 2009. PMID: 19491936 Review.
-
Folding up and Moving on-Nascent Protein Folding on the Ribosome.J Mol Biol. 2018 Oct 26;430(22):4580-4591. doi: 10.1016/j.jmb.2018.06.050. Epub 2018 Jul 5. J Mol Biol. 2018. PMID: 29981746 Free PMC article. Review.
Cited by
-
Mapping Protein-Protein Interactions at Birth: Single-Particle Cryo-EM Analysis of a Ribosome-Nascent Globin Complex.ACS Cent Sci. 2024 Feb 1;10(2):385-401. doi: 10.1021/acscentsci.3c00777. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435509 Free PMC article.
-
Nascent chains derived from a foldable protein sequence interact with specific ribosomal surface sites near the exit tunnel.Sci Rep. 2024 May 29;14(1):12324. doi: 10.1038/s41598-024-61274-1. Sci Rep. 2024. PMID: 38811604 Free PMC article.
-
Fluorescence Anisotropy for Detailed Analysis of Doxorubicin Loading into DNA Origami Nanocarriers for Drug Delivery.ACS Appl Nano Mater. 2025 Jun 24;8(26):13274-13284. doi: 10.1021/acsanm.5c01518. eCollection 2025 Jul 4. ACS Appl Nano Mater. 2025. PMID: 40636881 Free PMC article.
-
Protein folding in the cell: challenges and progress.Curr Opin Struct Biol. 2011 Feb;21(1):32-41. doi: 10.1016/j.sbi.2010.11.001. Epub 2010 Nov 26. Curr Opin Struct Biol. 2011. PMID: 21112769 Free PMC article. Review.
-
Unraveling co-translational protein folding: Concepts and methods.Methods. 2018 Mar 15;137:71-81. doi: 10.1016/j.ymeth.2017.11.007. Epub 2017 Dec 6. Methods. 2018. PMID: 29221924 Free PMC article. Review.
References
-
- Radford SE. Protein folding: progress made and promises ahead. Trends Biochem Sci. 2000;25:611–618. - PubMed
-
- Ellis RJ. The “bio” in biochemistry: Protein folding inside and outside the cell. Science. 1996;272:1448–1449. - PubMed
-
- Evans MS, Clark TF, Clark PL. Conformations of co-translational folding intermediates. Protein Pept Lett. 2005;12:189–195. - PubMed
-
- Bartlett AI, Radford SE. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat Struct Mol Biol. 2009;16:582–588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

