A specific docking site for DNA polymerase {alpha}-primase on the SV40 helicase is required for viral primosome activity, but helicase activity is dispensable
- PMID: 20685648
- PMCID: PMC2963361
- DOI: 10.1074/jbc.M110.156240
A specific docking site for DNA polymerase {alpha}-primase on the SV40 helicase is required for viral primosome activity, but helicase activity is dispensable
Abstract
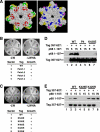
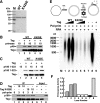
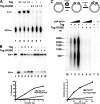
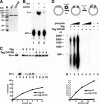
Replication of simian virus 40 (SV40) DNA, a model for eukaryotic chromosomal replication, can be reconstituted in vitro using the viral helicase (large tumor antigen, or Tag) and purified human proteins. Tag interacts physically with two cellular proteins, replication protein A and DNA polymerase α-primase (pol-prim), constituting the viral primosome. Like the well characterized primosomes of phages T7 and T4, this trio of proteins coordinates parental DNA unwinding with primer synthesis to initiate the leading strand at the viral origin and each Okazaki fragment on the lagging strand template. We recently determined the structure of a previously unrecognized pol-prim domain (p68N) that docks on Tag, identified the p68N surface that contacts Tag, and demonstrated its vital role in primosome function. Here, we identify the p68N-docking site on Tag by using structure-guided mutagenesis of the Tag helicase surface. A charge reverse substitution in Tag disrupted both p68N-binding and primosome activity but did not affect docking with other pol-prim subunits. Unexpectedly, the substitution also disrupted Tag ATPase and helicase activity, suggesting a potential link between p68N docking and ATPase activity. To assess this possibility, we examined the primosome activity of Tag with a single residue substitution in the Walker B motif. Although this substitution abolished ATPase and helicase activity as expected, it did not reduce pol-prim docking on Tag or primosome activity on single-stranded DNA, indicating that Tag ATPase is dispensable for primosome activity in vitro.
Figures


Similar articles
-
Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome.J Biol Chem. 2010 May 28;285(22):17112-22. doi: 10.1074/jbc.M110.116830. Epub 2010 Mar 16. J Biol Chem. 2010. PMID: 20234039 Free PMC article.
-
Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system.J Biol Chem. 2000 Jun 9;275(23):17328-37. doi: 10.1074/jbc.M000717200. J Biol Chem. 2000. PMID: 10747950
-
Mutational analysis of simian virus 40 T-antigen primosome activities in viral DNA replication.J Virol. 2002 May;76(10):5121-30. doi: 10.1128/jvi.76.10.5121-5130.2002. J Virol. 2002. PMID: 11967327 Free PMC article.
-
Choreography of bacteriophage T7 DNA replication.Curr Opin Chem Biol. 2011 Oct;15(5):580-6. doi: 10.1016/j.cbpa.2011.07.024. Epub 2011 Sep 9. Curr Opin Chem Biol. 2011. PMID: 21907611 Free PMC article. Review.
-
The initiation of simian virus 40 DNA replication in vitro.Crit Rev Biochem Mol Biol. 1997;32(6):503-68. doi: 10.3109/10409239709082001. Crit Rev Biochem Mol Biol. 1997. PMID: 9444478 Review.
Cited by
-
SV40 T antigen helicase domain regions responsible for oligomerisation regulate Okazaki fragment synthesis initiation.FEBS Open Bio. 2022 Mar;12(3):649-663. doi: 10.1002/2211-5463.13373. Epub 2022 Feb 7. FEBS Open Bio. 2022. PMID: 35073603 Free PMC article.
-
Origin DNA melting and unwinding in DNA replication.Curr Opin Struct Biol. 2010 Dec;20(6):756-62. doi: 10.1016/j.sbi.2010.08.009. Epub 2010 Oct 1. Curr Opin Struct Biol. 2010. PMID: 20870402 Free PMC article. Review.
-
Targeted chromosomal Escherichia coli:dnaB exterior surface residues regulate DNA helicase behavior to maintain genomic stability and organismal fitness.PLoS Genet. 2021 Nov 12;17(11):e1009886. doi: 10.1371/journal.pgen.1009886. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34767550 Free PMC article.
-
The conserved core enzymatic activities and the distinct dynamics of polyomavirus large T antigens.Arch Biochem Biophys. 2015 May 1;573:23-31. doi: 10.1016/j.abb.2015.02.033. Epub 2015 Mar 6. Arch Biochem Biophys. 2015. PMID: 25752954 Free PMC article.
-
Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress.J Biol Chem. 2012 Feb 24;287(9):6469-81. doi: 10.1074/jbc.M111.324582. Epub 2011 Dec 21. J Biol Chem. 2012. PMID: 22194613 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

