Information processing by biochemical networks: a dynamic approach
- PMID: 20685691
- PMCID: PMC3033026
- DOI: 10.1098/rsif.2010.0287
Information processing by biochemical networks: a dynamic approach
Abstract
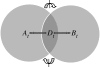
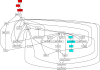
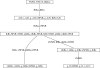
Understanding how information is encoded and transferred by biochemical networks is of fundamental importance in cellular and systems biology. This requires analysis of the relationships between the stochastic trajectories of the constituent molecular (or submolecular) species that comprise the network. We describe how to identify conditional independences between the trajectories or time courses of groups of species. These are robust network properties that provide important insight into how information is processed. An entire network can then be decomposed exactly into modules on informational grounds. In the context of signalling networks with multiple inputs, the approach identifies the routes and species involved in sequential information processing between input and output modules. An algorithm is developed which allows automated identification of decompositions for large networks and visualization using a tree that encodes the conditional independences. Only stoichiometric information is used and neither simulations nor knowledge of rate parameters are required. A bespoke version of the algorithm for signalling networks identifies the routes of sequential encoding between inputs and outputs, visualized as paths in the tree. Application to the toll-like receptor signalling network reveals that inputs can be informative in ways unanticipated by steady-state analyses, that the information processing structure is not well described as a bow tie, and that encoding for the interferon response is unusually sparse compared with other outputs of this innate immune system.
Figures



Similar articles
-
Identifying sources of variation and the flow of information in biochemical networks.Proc Natl Acad Sci U S A. 2012 May 15;109(20):E1320-8. doi: 10.1073/pnas.1119407109. Epub 2012 Apr 23. Proc Natl Acad Sci U S A. 2012. PMID: 22529351 Free PMC article.
-
Automated analysis of information processing, kinetic independence and modular architecture in biochemical networks using MIDIA.Bioinformatics. 2011 Feb 15;27(4):584-6. doi: 10.1093/bioinformatics/btq694. Epub 2010 Dec 14. Bioinformatics. 2011. PMID: 21159624
-
Signal integration and information transfer in an allosterically regulated network.NPJ Syst Biol Appl. 2019 Jul 18;5:23. doi: 10.1038/s41540-019-0100-9. eCollection 2019. NPJ Syst Biol Appl. 2019. PMID: 31341635 Free PMC article.
-
Information theory and signal transduction systems: from molecular information processing to network inference.Semin Cell Dev Biol. 2014 Nov;35:98-108. doi: 10.1016/j.semcdb.2014.06.011. Epub 2014 Jun 19. Semin Cell Dev Biol. 2014. PMID: 24953199 Review.
-
Engineering Aspects of Olfaction.In: Persaud KC, Marco S, Gutiérrez-Gálvez A, editors. Neuromorphic Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2013. Chapter 1. In: Persaud KC, Marco S, Gutiérrez-Gálvez A, editors. Neuromorphic Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2013. Chapter 1. PMID: 26042329 Free Books & Documents. Review.
Cited by
-
Identifying sources of variation and the flow of information in biochemical networks.Proc Natl Acad Sci U S A. 2012 May 15;109(20):E1320-8. doi: 10.1073/pnas.1119407109. Epub 2012 Apr 23. Proc Natl Acad Sci U S A. 2012. PMID: 22529351 Free PMC article.
-
Information transfer by leaky, heterogeneous, protein kinase signaling systems.Proc Natl Acad Sci U S A. 2014 Jan 21;111(3):E326-33. doi: 10.1073/pnas.1314446111. Epub 2014 Jan 6. Proc Natl Acad Sci U S A. 2014. PMID: 24395805 Free PMC article.
-
Carving Nature at Its Joints: A Comparison of CEMI Field Theory with Integrated Information Theory and Global Workspace Theory.Entropy (Basel). 2023 Dec 8;25(12):1635. doi: 10.3390/e25121635. Entropy (Basel). 2023. PMID: 38136515 Free PMC article.
-
Cutting the wires: modularization of cellular networks for experimental design.Biophys J. 2014 Jan 7;106(1):321-31. doi: 10.1016/j.bpj.2013.11.2960. Biophys J. 2014. PMID: 24411264 Free PMC article.
-
The fidelity of dynamic signaling by noisy biomolecular networks.PLoS Comput Biol. 2013;9(3):e1002965. doi: 10.1371/journal.pcbi.1002965. Epub 2013 Mar 28. PLoS Comput Biol. 2013. PMID: 23555208 Free PMC article.
References
-
- Barkai N., Shilo B.-Z. 2007. Variability and robustness in biomolecular systems. Mol. Cell 28, 755–76010.1016/j.molcel.2007.11.013 (doi:10.1016/j.molcel.2007.11.013) - DOI - DOI - PubMed
-
- Nurse P. 2008. Life, logic and information. Nature 425, 424–42610.1038/454424a (doi:10.1038/454424a) - DOI - DOI - PubMed
-
- Samoilov M., Arkin A., Ross J. 2002. Signal processing by simple chemical systems. J. Phys. Chem. A 106, 10 205–10 22110.1021/jp025846z (doi:10.1021/jp025846z) - DOI - DOI
-
- Shankaran H., Ippolito D. L., Chrisler W. B., Resat H., Bollinger N., Opresko L. K., Wiley H. S. 2009. Rapid and sustained nuclear–cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 5, 332.10.1038/msb.2009.90 (doi:10.1038/msb.2009.90) - DOI - DOI - PMC - PubMed
-
- Berridge M. J., Bootman M. D., Roderick H. L. 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–52910.1038/nrm1155 (doi:10.1038/nrm1155) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

