Using protein design algorithms to understand the molecular basis of disease caused by protein-DNA interactions: the Pax6 example
- PMID: 20685816
- PMCID: PMC2995082
- DOI: 10.1093/nar/gkq683
Using protein design algorithms to understand the molecular basis of disease caused by protein-DNA interactions: the Pax6 example
Abstract
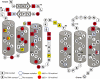
Quite often a single or a combination of protein mutations is linked to specific diseases. However, distinguishing from sequence information which mutations have real effects in the protein's function is not trivial. Protein design tools are commonly used to explain mutations that affect protein stability, or protein-protein interaction, but not for mutations that could affect protein-DNA binding. Here, we used the protein design algorithm FoldX to model all known missense mutations in the paired box domain of Pax6, a highly conserved transcription factor involved in eye development and in several diseases such as aniridia. The validity of FoldX to deal with protein-DNA interactions was demonstrated by showing that high levels of accuracy can be achieved for mutations affecting these interactions. Also we showed that protein-design algorithms can accurately reproduce experimental DNA-binding logos. We conclude that 88% of the Pax6 mutations can be linked to changes in intrinsic stability (77%) and/or to its capabilities to bind DNA (30%). Our study emphasizes the importance of structure-based analysis to understand the molecular basis of diseases and shows that protein-DNA interactions can be analyzed to the same level of accuracy as protein stability, or protein-protein interactions.
Figures



Similar articles
-
Functional analysis of missense mutations G36A and G51A in PAX6, and PAX6(5a) causing ocular anomalies.Exp Eye Res. 2011 Jul;93(1):40-9. doi: 10.1016/j.exer.2011.04.001. Epub 2011 Apr 19. Exp Eye Res. 2011. PMID: 21524647
-
Identification of dominant FOXE3 and PAX6 mutations in patients with congenital cataract and aniridia.Mol Vis. 2010 Aug 22;16:1705-11. Mol Vis. 2010. PMID: 20806047 Free PMC article.
-
Gene regulation by PAX6: structural-functional correlations of missense mutants and transcriptional control of Trpm3/miR-204.Mol Vis. 2014 Mar 6;20:270-82. eCollection 2014. Mol Vis. 2014. PMID: 24623969 Free PMC article.
-
Aniridia with a heterozygous PAX6 mutation in which the pituitary function was partially impaired.Intern Med. 2014;53(1):39-42. doi: 10.2169/internalmedicine.53.1184. Intern Med. 2014. PMID: 24390526 Review.
-
FoldX as Protein Engineering Tool: Better Than Random Based Approaches?Comput Struct Biotechnol J. 2018 Feb 3;16:25-33. doi: 10.1016/j.csbj.2018.01.002. eCollection 2018. Comput Struct Biotechnol J. 2018. PMID: 30275935 Free PMC article. Review.
Cited by
-
Congenital Adrenal Hyperplasia (CAH) due to 21-Hydroxylase Deficiency: A Comprehensive Focus on 233 Pathogenic Variants of CYP21A2 Gene.Mol Diagn Ther. 2018 Jun;22(3):261-280. doi: 10.1007/s40291-018-0319-y. Mol Diagn Ther. 2018. PMID: 29450859 Review.
-
Assessing the correlation between mutant rhodopsin stability and the severity of retinitis pigmentosa.Mol Vis. 2014 Feb 7;20:183-99. eCollection 2014. Mol Vis. 2014. PMID: 24520188 Free PMC article.
-
UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions.Nucleic Acids Res. 2015 Jan;43(Database issue):D117-22. doi: 10.1093/nar/gku1045. Epub 2014 Nov 5. Nucleic Acids Res. 2015. PMID: 25378322 Free PMC article.
-
Structure-based activity prediction of CYP21A2 stability variants: A survey of available gene variations.Sci Rep. 2016 Dec 14;6:39082. doi: 10.1038/srep39082. Sci Rep. 2016. PMID: 27966633 Free PMC article.
-
Atomistic modeling of protein-DNA interaction specificity: progress and applications.Curr Opin Struct Biol. 2012 Aug;22(4):397-405. doi: 10.1016/j.sbi.2012.06.002. Epub 2012 Jul 13. Curr Opin Struct Biol. 2012. PMID: 22796087 Free PMC article. Review.
References
-
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. - PubMed
-
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. - PubMed
-
- Tsonis PA, Fuentes EJ. Focus on molecules: Pax-6, the eye master. Exp. Eye Res. 2006;83:233–234. - PubMed
-
- van Heyningen V, Williamson KA. PAX6 in sensory development. Hum. Mol. Genet. 2002;11:1161–1167. - PubMed
-
- Kokotas H, Petersen MB. Clinical and molecular aspects of aniridia. Clin. Genet. 2010;77:409–420. - PubMed

