The human papillomavirus type 16 E5 oncoprotein inhibits epidermal growth factor trafficking independently of endosome acidification
- PMID: 20686024
- PMCID: PMC2950579
- DOI: 10.1128/JVI.00831-10
The human papillomavirus type 16 E5 oncoprotein inhibits epidermal growth factor trafficking independently of endosome acidification
Abstract
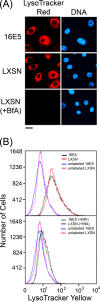
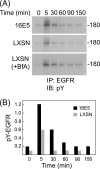
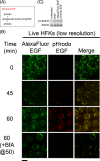
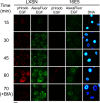
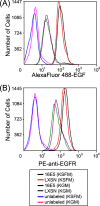
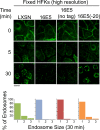
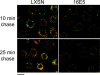
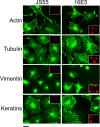
The human papillomavirus type 16 E5 oncoprotein (16E5) enhances acute, ligand-dependent activation of the epidermal growth factor receptor (EGFR) and concomitantly alkalinizes endosomes, presumably by binding to the 16-kDa "c" subunit of the V-ATPase proton pump (16K) and inhibiting V-ATPase function. However, the relationship between 16K binding, endosome alkalinization, and altered EGFR signaling remains unclear. Using an antibody that we generated against 16K, we found that 16E5 associated with only a small fraction of endogenous 16K in keratinocytes, suggesting that it was unlikely that E5 could significantly affect V-ATPase function by direct inhibition. Nevertheless, E5 inhibited the acidification of endosomes, as determined by a new assay using a biologically active, pH-sensitive fluorescent EGF conjugate. Since we also found that 16E5 did not alter cell surface EGF binding, the number of EGFRs on the cell surface, or the endocytosis of prebound EGF, we postulated that it might be blocking the fusion of early endosomes with acidified vesicles. Our studies with pH-sensitive and -insensitive fluorescent EGF conjugates and fluorescent dextran confirmed that E5 prevented endosome maturation (acidification and enlargement) by inhibiting endosome fusion. The E5-dependent defect in vesicle fusion was not due to detectable disruption of actin, tubulin, vimentin, or cytokeratin filaments, suggesting that membrane fusion was being directly affected rather than vesicle transport. Perhaps most importantly, while bafilomycin A(1) (like E5) binds to 16K and inhibits endosome acidification, it did not mimic the ability of E5 to inhibit endosome enlargement or the trafficking of EGF. Thus, 16E5 alters EGF endocytic trafficking via a pH-independent inhibition of vesicle fusion.
Figures

Similar articles
-
The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes.J Virol. 1995 May;69(5):3185-92. doi: 10.1128/JVI.69.5.3185-3192.1995. J Virol. 1995. PMID: 7707548 Free PMC article.
-
Binding of human papillomavirus 16 E5 to the 16 kDa subunit c (proteolipid) of the vacuolar H+-ATPase can be dissociated from the E5-mediated epidermal growth factor receptor overactivation.Oncogene. 2000 Aug 3;19(33):3727-32. doi: 10.1038/sj.onc.1203718. Oncogene. 2000. PMID: 10949926
-
The human papillomavirus type 16 E5 oncoprotein synergizes with EGF-receptor signaling to enhance cell cycle progression and the down-regulation of p27(Kip1).Virology. 2010 Apr 25;400(1):44-52. doi: 10.1016/j.virol.2010.01.009. Epub 2010 Feb 7. Virology. 2010. PMID: 20144468
-
Regulation of epithelial growth factor receptors by the oncoprotein E5 during the HPV16 differentiation-dependent life cycle.Tumour Virus Res. 2025 Jun;19:200315. doi: 10.1016/j.tvr.2025.200315. Epub 2025 Mar 7. Tumour Virus Res. 2025. PMID: 40057277 Free PMC article. Review.
-
Mechanisms of cell transformation by papillomavirus E5 proteins.Oncogene. 2001 Nov 26;20(54):7866-73. doi: 10.1038/sj.onc.1204915. Oncogene. 2001. PMID: 11753669 Review.
Cited by
-
Conditional cell reprogramming involves non-canonical β-catenin activation and mTOR-mediated inactivation of Akt.PLoS One. 2017 Jul 10;12(7):e0180897. doi: 10.1371/journal.pone.0180897. eCollection 2017. PLoS One. 2017. PMID: 28700668 Free PMC article.
-
High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors.J Virol. 2012 May;86(9):5341-51. doi: 10.1128/JVI.06243-11. Epub 2012 Feb 22. J Virol. 2012. PMID: 22357280 Free PMC article.
-
Targeting HER (ERBB) signaling in head and neck cancer: An essential update.Mol Aspects Med. 2015 Nov;45:74-86. doi: 10.1016/j.mam.2015.07.001. Epub 2015 Jul 7. Mol Aspects Med. 2015. PMID: 26163475 Free PMC article. Review.
-
Development of Novel Single-Chain Antibodies against the Hydrophobic HPV-16 E5 Protein.Biomed Res Int. 2018 Jun 20;2018:5809028. doi: 10.1155/2018/5809028. eCollection 2018. Biomed Res Int. 2018. PMID: 30027096 Free PMC article.
-
The human papillomavirus oncoproteins: a review of the host pathways targeted on the road to transformation.J Gen Virol. 2021 Mar;102(3):001540. doi: 10.1099/jgv.0.001540. Epub 2021 Jan 11. J Gen Virol. 2021. PMID: 33427604 Free PMC article. Review.
References
-
- Adam, J. L., M. W. Briggs, and D. J. McCance. 2000. A mutagenic analysis of the E5 protein of human papillomavirus type 16 reveals that E5 binding to the vacuolar H+-ATPase is not sufficient for biological activity, using mammalian and yeast expression systems. Virology 272:315-325. - PubMed
-
- Andresson, T., J. Sparkowski, D. J. Goldstein, and R. Schlegel. 1995. Vacuolar H+-ATPase mutants transform cells and define a binding site for the papillomavirus E5 oncoprotein. J. Biol. Chem. 270:6830-6837. - PubMed
-
- Ashby, A. D., L. Meagher, M. S. Campo, and M. E. Finbow. 2001. E5 transforming proteins of papillomaviruses do not disturb the activity of the vacuolar H+-ATPase. J. Gen. Virol. 82:2353-2362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

