Convergence of Kaposi's sarcoma-associated herpesvirus reactivation with Epstein-Barr virus latency and cellular growth mediated by the notch signaling pathway in coinfected cells
- PMID: 20686042
- PMCID: PMC2950570
- DOI: 10.1128/JVI.00894-10
Convergence of Kaposi's sarcoma-associated herpesvirus reactivation with Epstein-Barr virus latency and cellular growth mediated by the notch signaling pathway in coinfected cells
Abstract
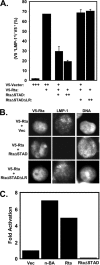
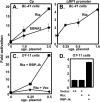
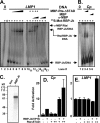
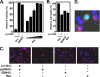
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent of primary effusion lymphoma (PEL). All PEL cell lines are infected with KSHV, and 70% are coinfected with Epstein-Barr virus (EBV). KSHV reactivation from latency requires promoter-specific transactivation by the KSHV Rta protein through interactions with RBP-Jk (CSL), the cellular DNA-binding component of the Notch signal transduction pathway. EBV transformation of primary B cells requires EBV nuclear antigen 2 (EBNA-2) to interact with RBP-Jk to direct the latent viral and cellular gene expression program. Although KSHV Rta and EBV EBNA-2 both require RBP-Jk for transactivation, previous studies have suggested that RBP-Jk-dependent transactivators do not function identically. We have found that the EBV latent protein LMP-1 is expressed in less than 5% of KSHV(+)/EBV(+) PEL cells but is induced in an Rta-dependent fashion when KSHV reactivates. KSHV Rta transactivates the EBV latency promoters in an RBP-Jk-dependent fashion and forms a ternary complex with RBP-Jk on the promoters. In B cells that are conditionally transformed by EBV alone, we show that KSHV Rta complements a short-term EBNA-2 growth deficiency in an autocrine/paracrine manner. Complementation of EBNA-2 deficiency by Rta depends on RBP-Jk and LMP-1, and Rta transactivation is required for optimal growth of KSHV(+)/EBV(+) PEL lines. Our data suggest that Rta can contribute to EBV-driven cellular growth by transactivating RBP-Jk-dependent EBV latency genes. However, our data also suggest that EBNA-2 and Rta induce distinct alterations in the cellular proteomes that contribute to the growth of infected cells.
Figures




Similar articles
-
The cellular Notch1 protein promotes KSHV reactivation in an Rta-dependent manner.J Virol. 2024 Aug 20;98(8):e0078824. doi: 10.1128/jvi.00788-24. Epub 2024 Jul 8. J Virol. 2024. PMID: 38975769 Free PMC article.
-
Kaposi's Sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway.J Virol. 2006 Oct;80(19):9697-709. doi: 10.1128/JVI.00746-06. J Virol. 2006. PMID: 16973574 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus Rta tetramers make high-affinity interactions with repetitive DNA elements in the Mta promoter to stimulate DNA binding of RBP-Jk/CSL.J Virol. 2011 Nov;85(22):11901-15. doi: 10.1128/JVI.05479-11. Epub 2011 Aug 31. J Virol. 2011. PMID: 21880753 Free PMC article.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
-
KSHV and the Role of Notch Receptor Dysregulation in Disease Progression.Pathogens. 2017 Aug 4;6(3):34. doi: 10.3390/pathogens6030034. Pathogens. 2017. PMID: 28777778 Free PMC article. Review.
Cited by
-
Kaposi Sarcoma-Associated Herpesvirus Infection and Endemic Burkitt Lymphoma.J Infect Dis. 2020 Jun 16;222(1):111-120. doi: 10.1093/infdis/jiaa060. J Infect Dis. 2020. PMID: 32072172 Free PMC article.
-
Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes.BMC Genomics. 2011 Dec 20;12:625. doi: 10.1186/1471-2164-12-625. BMC Genomics. 2011. PMID: 22185355 Free PMC article.
-
Analysis of the ORFK1 hypervariable regions reveal distinct HHV-8 clustering in Kaposi's sarcoma and non-Kaposi's cases.J Exp Clin Cancer Res. 2015 Jan 16;34(1):1. doi: 10.1186/s13046-014-0119-0. J Exp Clin Cancer Res. 2015. PMID: 25592960 Free PMC article.
-
KSHV Rta Promoter Specification and Viral Reactivation.Front Microbiol. 2012 Feb 14;3:30. doi: 10.3389/fmicb.2012.00030. eCollection 2012. Front Microbiol. 2012. PMID: 22347875 Free PMC article.
-
The Role of Coinfections in the EBV-Host Broken Equilibrium.Viruses. 2021 Jul 19;13(7):1399. doi: 10.3390/v13071399. Viruses. 2021. PMID: 34372605 Free PMC article. Review.
References
-
- Abramoff, M., P. Magelhaes, and S. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42.
-
- Adriaenssens, E., A. Mougel, G. Goormachtigh, E. Loing, V. Fafeur, C. Auriault, and J. Coll. 2004. A novel dominant-negative mutant form of Epstein-Barr virus latent membrane protein-1 (LMP1) selectively and differentially impairs LMP1 and TNF signaling pathways. Oncogene 23:2681-2693. - PubMed
-
- Brennan, P., J. E. Floettmann, A. Mehl, M. Jones, and M. Rowe. 2001. Mechanism of action of a novel latent membrane protein-1 dominant negative. J. Biol. Chem. 276:1195-1203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

