Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases
- PMID: 20686113
- PMCID: PMC2930558
- DOI: 10.1073/pnas.1009424107
Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases
Abstract
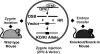
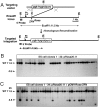
Gene targeting by homologous recombination in embryonic stem cells is extensively used to generate specific mouse mutants. However, most mammalian species lack tools for targeted gene manipulation. Since double-strand breaks strongly increase the rate of homologous recombination at genomic loci, we explored whether gene targeting can be directly performed in zygotes by the use of zinc-finger nucleases. Here we report that gene targeting is achieved in 1.7-4.5% of murine one-cell embryos upon the coinjection of targeting vectors with zinc-finger nucleases, without preselection. These findings enable the manipulation of the mammalian germ line in a single step in zygotes, independent of ES cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. - PubMed
-
- McCreath KJ, et al. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. - PubMed
-
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

