Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA
- PMID: 20686355
- PMCID: PMC2928650
- DOI: 10.4161/cc.9.15.12257
Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA
Abstract
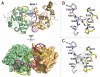
Ets1 is a member of the Ets family of transcription factors. Ets1 is autoinhibited and its activation requires heterodimerization with a partner protein or DNA-mediated homodimerization for cooperative DNA binding. In the latter case, Ets1 molecules bind to palindromic sequences in which two Ets-binding sites (EBS) are separated by four base pairs, for example in the promoters of stromelysin-1 and p53. Interestingly, counteraction of autoinhibition requires the autoinhibitory region encoded by exon VII of the gene. The structural basis for the requirement of autoinhibitory sequences for Ets1 binding to palindromic EBS still remains unresolved. Here we report the crystal structure of two Ets1 molecules bound to an EBS palindrome of the stromelysin-1 promoter DNA, providing a plausible explanation for the requirement of exon VII-encoded sequences for Ets1 cooperative DNA binding. The proposed mechanism was verified both in vitro by surface plasmon resonance and in vivo by transcription-based assays.
Figures





References
-
- Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. - PubMed
-
- Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 2006;54:149–163. - PubMed
-
- Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Secondary structure of the ETS domain places murine Ets-1 in the superfamily of winged helix-turn-helix DNA-binding proteins. Biochemistry. 1994;33:13509–13516. - PubMed
-
- Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. - PubMed
-
- Leprince D, Gegonne A, Coll J, de Taisne C, Schneeberger A, Lagrou C, et al. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983;306:395–397. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
