SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome
- PMID: 20686364
- PMCID: PMC3093916
- DOI: 10.4161/cbt.10.8.12914
SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome
Abstract
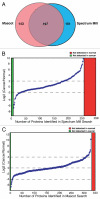
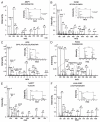
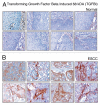
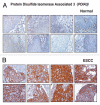
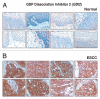
The identification of secreted proteins that are differentially expressed between non-neoplastic and esophageal squamous cell carcinoma (ESCC) cells can provide potential biomarkers of ESCC. We used a SILAC-based quantitative proteomic approach to compare the secretome of ESCC cells with that of non-neoplastic esophageal squamous epithelial cells. Proteins were resolved by SDS-PAGE, and tandem mass spectrometry analysis (LC-MS/MS) of in-gel trypsin-digested peptides was carried out on a high-accuracy qTOF mass spectrometer. In total, we identified 441 proteins in the combined secretomes, including 120 proteins with > 2-fold upregulation in the ESCC secretome vs. that of non-neoplastic esophageal squamous epithelial cells. In this study, several potential protein biomarkers previously known to be increased in ESCC including matrix metalloproteinase 1, transferrin receptor, and transforming growth factor beta-induced 68 kDa were identified as overexpressed in the ESCC-derived secretome. In addition, we identified several novel proteins that have not been previously reported to be associated with ESCC. Among the novel candidate proteins identified, protein disulfide isomerase family a member 3 (PDIA3), GDP dissociation inhibitor 2 (GDI2), and lectin galactoside binding soluble 3 binding protein (LGALS3BP) were further validated by immunoblot analysis and immunohistochemical labeling using tissue microarrays. This tissue microarray analysis showed overexpression of protein disulfide isomerase family a member 3, GDP dissociation inhibitor 2, and lectin galactoside binding soluble 3 binding protein in 93%, 93% and 87% of 137 ESCC cases, respectively. Hence, we conclude that these potential biomarkers are excellent candidates for further evaluation to test their role and efficacy in the early detection of ESCC.
Figures



Comment in
-
Maximizing early detection of esophageal squamous cell carcinoma via SILAC-proteomics.Cancer Biol Ther. 2010 Oct 15;10(8):811-3. doi: 10.4161/cbt.10.8.13754. Epub 2010 Oct 15. Cancer Biol Ther. 2010. PMID: 20953140 Free PMC article. No abstract available.
Similar articles
-
Maximizing early detection of esophageal squamous cell carcinoma via SILAC-proteomics.Cancer Biol Ther. 2010 Oct 15;10(8):811-3. doi: 10.4161/cbt.10.8.13754. Epub 2010 Oct 15. Cancer Biol Ther. 2010. PMID: 20953140 Free PMC article. No abstract available.
-
Quantitative tissue proteomics of esophageal squamous cell carcinoma for novel biomarker discovery.Cancer Biol Ther. 2011 Sep 15;12(6):510-22. doi: 10.4161/cbt.12.6.16833. Epub 2011 Sep 15. Cancer Biol Ther. 2011. PMID: 21743296 Free PMC article.
-
Identification of galectin-7 as a potential biomarker for esophageal squamous cell carcinoma by proteomic analysis.BMC Cancer. 2010 Jun 15;10:290. doi: 10.1186/1471-2407-10-290. BMC Cancer. 2010. PMID: 20546628 Free PMC article.
-
An overview of esophageal squamous cell carcinoma proteomics.J Proteomics. 2012 Jun 18;75(11):3129-37. doi: 10.1016/j.jprot.2012.04.025. Epub 2012 Apr 27. J Proteomics. 2012. PMID: 22564818 Review.
-
Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes.Proteomics. 2012 Feb;12(4-5):666-72. doi: 10.1002/pmic.201100488. Epub 2012 Jan 19. Proteomics. 2012. PMID: 22246955 Review.
Cited by
-
Mass spectrometry-based proteomics in molecular diagnostics: discovery of cancer biomarkers using tissue culture.Biomed Res Int. 2013;2013:783131. doi: 10.1155/2013/783131. Epub 2013 Mar 17. Biomed Res Int. 2013. PMID: 23586059 Free PMC article. Review.
-
SILAC-based quantitative MS approach for real-time recording protein-mediated cell-cell interactions.Sci Rep. 2018 May 31;8(1):8441. doi: 10.1038/s41598-018-26262-2. Sci Rep. 2018. PMID: 29855483 Free PMC article.
-
A Sensitized Screen for Genes Promoting Invadopodia Function In Vivo: CDC-42 and Rab GDI-1 Direct Distinct Aspects of Invadopodia Formation.PLoS Genet. 2016 Jan 14;12(1):e1005786. doi: 10.1371/journal.pgen.1005786. eCollection 2016 Jan. PLoS Genet. 2016. PMID: 26765257 Free PMC article.
-
Methodologies to decipher the cell secretome.Biochim Biophys Acta. 2013 Nov;1834(11):2226-32. doi: 10.1016/j.bbapap.2013.01.022. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376189 Free PMC article. Review.
-
Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma.Mol Cancer. 2018 Feb 19;17(1):54. doi: 10.1186/s12943-018-0790-4. Mol Cancer. 2018. PMID: 29455652 Free PMC article. Review.
References
-
- Oka D, Yamashita S, Tomioka T, Nakanishi Y, Kato H, Kaminishi M, Ushijima T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–3426. - PubMed
-
- Yu C, Chen K, Zheng H, Guo X, Jia W, Li M, Zeng M, Li J, Song L. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
