Analysis of human and animal fecal microbiota for microbial source tracking
- PMID: 20686512
- PMCID: PMC3105695
- DOI: 10.1038/ismej.2010.120
Analysis of human and animal fecal microbiota for microbial source tracking
Abstract
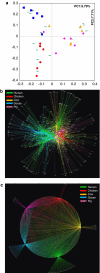
Microbial compositions of human and animal feces from South Korea were analyzed and characterized. In total, 38 fecal samples (14 healthy adult humans, 6 chickens, 6 cows, 6 pigs and 6 geese) were analyzed by 454 pyrosequencing of the V2 region of the 16S rRNA gene. Four major phyla, Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes, were identified in the samples. Principal coordinate analysis suggested that microbiota from the same host species generally clustered, with the exception of those from humans, which exhibited sample-specific compositions. A network-based analysis revealed that several operational taxonomic units (OTUs), such as Lactobacillus sp., Clostridium sp. and Prevotella sp., were commonly identified in all fecal sources. Other OTUs were present only in fecal samples from a single organism. For example, Yania sp. and Bifidobacterium sp. were identified specifically in chicken and human fecal samples, respectively. These specific OTUs or their respective biological markers could be useful for identifying the sources of fecal contamination in water by microbial source tracking.
Figures
References
-
- Byappanahalli MN, Przybyla-Kelly K, Shively DA, Whitman RL. Environmental occurrence of the enterococcal surface protein (esp) gene is an unreliable indicator of human fecal contamination. Environ Sci Technol. 2008;42:8014–8020. - PubMed
-
- Dorai-Raj S, Grady JO, Colleran E. Specificity and sensitivity evaluation of novel and existing Bacteroidales and Bifidobacteria-specific PCR assays on feces and sewage samples and their application for microbial source tracking in Ireland. Water Res. 2009;43:4980–4988. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

