Common ADRB2 haplotypes derived from 26 polymorphic sites direct beta2-adrenergic receptor expression and regulation phenotypes
- PMID: 20686604
- PMCID: PMC2912278
- DOI: 10.1371/journal.pone.0011819
Common ADRB2 haplotypes derived from 26 polymorphic sites direct beta2-adrenergic receptor expression and regulation phenotypes
Abstract
Background: The beta2-adrenergic receptor (beta2AR) is expressed on numerous cell-types including airway smooth muscle cells and cardiomyocytes. Drugs (agonists or antagonists) acting at these receptors for treatment of asthma, chronic obstructive pulmonary disease, and heart failure show substantial interindividual variability in response. The ADRB2 gene is polymorphic in noncoding and coding regions, but virtually all ADRB2 association studies have utilized the two common nonsynonymous coding SNPs, often reaching discrepant conclusions.
Methodology/principal findings: We constructed the 8 common ADRB2 haplotypes derived from 26 polymorphisms in the promoter, 5'UTR, coding, and 3'UTR of the intronless ADRB2 gene. These were cloned into an expression construct lacking a vector-based promoter, so that beta2AR expression was driven by its promoter, and steady state expression could be modified by polymorphisms throughout ADRB2 within a haplotype. "Whole-gene" transfections were performed with COS-7 cells and revealed 4 haplotypes with increased cell surface beta2AR protein expression compared to the others. Agonist-promoted downregulation of beta2AR protein expression was also haplotype-dependent, and was found to be increased for 2 haplotypes. A phylogenetic tree of the haplotypes was derived and annotated by cellular phenotypes, revealing a pattern potentially driven by expression.
Conclusions/significance: Thus for obstructive lung disease, the initial bronchodilator response from intermittent administration of beta-agonist may be influenced by certain beta2AR haplotypes (expression phenotypes), while other haplotypes may influence tachyphylaxis during the response to chronic therapy (downregulation phenotypes). An ideal clinical outcome of high expression and less downregulation was found for two haplotypes. Haplotypes may also affect heart failure antagonist therapy, where beta2AR increase inotropy and are anti-apoptotic. The haplotype-specific expression and regulation phenotypes found in this transfection-based system suggest that the density of genetic information in the form of these haplotypes, or haplotype-clusters with similar phenotypes can potentially provide greater discrimination of phenotype in human disease and pharmacogenomic association studies.
Conflict of interest statement
Figures
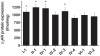





Similar articles
-
Effect of β2-adrenergic receptor gene (ADRB2) 3' untranslated region polymorphisms on inhaled corticosteroid/long-acting β2-adrenergic agonist response.Respir Res. 2012 May 4;13(1):37. doi: 10.1186/1465-9921-13-37. Respir Res. 2012. PMID: 22559839 Free PMC article. Clinical Trial.
-
Variable-length poly-C tract polymorphisms of the beta2-adrenergic receptor 3'-UTR alter expression and agonist regulation.Am J Physiol Lung Cell Mol Physiol. 2008 Feb;294(2):L190-5. doi: 10.1152/ajplung.00277.2007. Epub 2007 Nov 16. Am J Physiol Lung Cell Mol Physiol. 2008. PMID: 18024720 Free PMC article.
-
Polymorphisms of the 5' leader cistron of the human beta2-adrenergic receptor regulate receptor expression.J Clin Invest. 1998 Dec 1;102(11):1927-32. doi: 10.1172/JCI4862. J Clin Invest. 1998. PMID: 9835617 Free PMC article.
-
The significance of beta2-adrenergic receptor polymorphisms in asthma.Curr Opin Pulm Med. 2006 Jan;12(1):12-7. doi: 10.1097/01.mcp.0000198068.50457.95. Curr Opin Pulm Med. 2006. PMID: 16357573 Review.
-
Beta-2 adrenergic receptor genetic polymorphisms and asthma.J Clin Pharm Ther. 2009 Dec;34(6):631-43. doi: 10.1111/j.1365-2710.2009.01066.x. J Clin Pharm Ther. 2009. PMID: 20175796 Review.
Cited by
-
Effect of β2-adrenergic receptor gene (ADRB2) 3' untranslated region polymorphisms on inhaled corticosteroid/long-acting β2-adrenergic agonist response.Respir Res. 2012 May 4;13(1):37. doi: 10.1186/1465-9921-13-37. Respir Res. 2012. PMID: 22559839 Free PMC article. Clinical Trial.
-
Pharmacogenetic Factors Shaping Treatment Outcomes in Chronic Obstructive Pulmonary Disease.Genes (Basel). 2025 Mar 6;16(3):314. doi: 10.3390/genes16030314. Genes (Basel). 2025. PMID: 40149465 Free PMC article. Review.
-
ADRB2 polymorphisms predict the risk of myocardial infarction and coronary artery disease.Genet Mol Biol. 2015 Dec;38(4):433-43. doi: 10.1590/S1415-475738420140234. Epub 2015 Nov 24. Genet Mol Biol. 2015. PMID: 26692153 Free PMC article.
-
Correlation in gene expression between the aggravation of chronic obstructive pulmonary disease and the occurrence of complications.Bioengineered. 2020 Dec;11(1):1245-1257. doi: 10.1080/21655979.2020.1839216. Bioengineered. 2020. PMID: 33108241 Free PMC article.
-
MicroRNA let-7 establishes expression of beta2-adrenergic receptors and dynamically down-regulates agonist-promoted down-regulation.Proc Natl Acad Sci U S A. 2011 Apr 12;108(15):6246-51. doi: 10.1073/pnas.1101439108. Epub 2011 Mar 29. Proc Natl Acad Sci U S A. 2011. PMID: 21447718 Free PMC article.
References
-
- Barnes PJ. Beta-adrenergic receptors and their regulation. Am J Respir Crit Care Med. 1995;152:838–860. - PubMed
-
- Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–1070. - PubMed
-
- van Campen LC, Visser FC, Visser CA. Ejection fraction improvement by beta-blocker treatment in patients with heart failure: an analysis of studies published in the literature. J Cardiovasc Pharmacol. 1998;32(Suppl 1):S31–S35. - PubMed
-
- Reihsaus E, Innis M, MacIntyre N, Liggett SB. Mutations in the gene encoding for the β2-adrenergic receptor in normal and asthmatic subjects. Am J Resp Cell Mol Biol. 1993;8:334–339. - PubMed
-
- Hall IP, Liggett SB. Pharmacogenetics of respiratory disease. In: Hall IP, Pirmohamed M, editors. Pharmacogenetics. New York: Informa Healthcare; 2006. pp. 91–111.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

