Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient
- PMID: 20686659
- PMCID: PMC2912394
- DOI: 10.1371/journal.ppat.1001024
Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient
Abstract
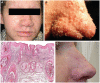
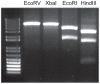
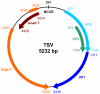
The Polyomaviridae constitute a family of small DNA viruses infecting a variety of hosts. In humans, polyomaviruses can cause infections of the central nervous system, urinary tract, skin, and possibly the respiratory tract. Here we report the identification of a new human polyomavirus in plucked facial spines of a heart transplant patient with trichodysplasia spinulosa, a rare skin disease exclusively seen in immunocompromized patients. The trichodysplasia spinulosa-associated polyomavirus (TSV) genome was amplified through rolling-circle amplification and consists of a 5232-nucleotide circular DNA organized similarly to known polyomaviruses. Two putative "early" (small and large T antigen) and three putative "late" (VP1, VP2, VP3) genes were identified. The TSV large T antigen contains several domains (e.g. J-domain) and motifs (e.g. HPDKGG, pRb family-binding, zinc finger) described for other polyomaviruses and potentially involved in cellular transformation. Phylogenetic analysis revealed a close relationship of TSV with the Bornean orangutan polyomavirus and, more distantly, the Merkel cell polyomavirus that is found integrated in Merkel cell carcinomas of the skin. The presence of TSV in the affected patient's skin was confirmed by newly designed quantitative TSV-specific PCR, indicative of a viral load of 10(5) copies per cell. After topical cidofovir treatment, the lesions largely resolved coinciding with a reduction in TSV load. PCR screening demonstrated a 4% prevalence of TSV in an unrelated group of immunosuppressed transplant recipients without apparent disease. In conclusion, a new human polyomavirus was discovered and identified as the possible cause of trichodysplasia spinulosa in immunocompromized patients. The presence of TSV also in clinically unaffected individuals suggests frequent virus transmission causing subclinical, probably latent infections. Further studies have to reveal the impact of TSV infection in relation to other populations and diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

