The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella
- PMID: 20686667
- PMCID: PMC2912647
- DOI: 10.1371/journal.ppat.1001025
The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella
Erratum in
-
Correction: The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella.PLoS Pathog. 2010 Aug 5;6(8):10.1371/annotation/df7e26bc-4c62-43b4-865f-a39274d98ab3. doi: 10.1371/annotation/df7e26bc-4c62-43b4-865f-a39274d98ab3. PLoS Pathog. 2010. PMID: 20700452 Free PMC article.
Abstract
Salmonella enterica serovar Typhimurium is a common food-borne pathogen that induces inflammatory diarrhea and invades intestinal epithelial cells using a type three secretion system (T3SS) encoded within Salmonella pathogenicity island 1 (SPI1). The genes encoding the SPI1 T3SS are tightly regulated by a network of interacting transcriptional regulators involving three coupled positive feedback loops. While the core architecture of the SPI1 gene circuit has been determined, the relative roles of these interacting regulators and associated feedback loops are still unknown. To determine the function of this circuit, we measured gene expression dynamics at both population and single-cell resolution in a number of SPI1 regulatory mutants. Using these data, we constructed a mathematical model of the SPI1 gene circuit. Analysis of the model predicted that the circuit serves two functions. The first is to place a threshold on SPI1 activation, ensuring that the genes encoding the T3SS are expressed only in response to the appropriate combination of environmental and cellular cues. The second is to amplify SPI1 gene expression. To experimentally test these predictions, we rewired the SPI1 genetic circuit by changing its regulatory architecture. This enabled us to directly test our predictions regarding the function of the circuit by varying the strength and dynamics of the activating signal. Collectively, our experimental and computational results enable us to deconstruct this complex circuit and determine the role of its individual components in regulating SPI1 gene expression dynamics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
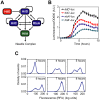


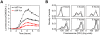
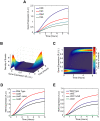
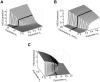
 and
and  . The parameter
. The parameter  specifies the degree by which the SPI1 regulators - HilC, HilD, and RtsA - can activate HilD expression, effectively the strength of positive feedback on HilD expression. The parameter
specifies the degree by which the SPI1 regulators - HilC, HilD, and RtsA - can activate HilD expression, effectively the strength of positive feedback on HilD expression. The parameter  specifies the strength of the signal activating HilD expression. (B) Effect of HilC and RtsA on HilD expression. Plot shows the steady-state concentration of HilD as a function of the parameters
specifies the strength of the signal activating HilD expression. (B) Effect of HilC and RtsA on HilD expression. Plot shows the steady-state concentration of HilD as a function of the parameters  ,
,  , and
, and  . The parameters
. The parameters  and
and  specify the degree by which the SPI1 regulators - HilC, HilD, and RtsA - can activate HilC and RtsA expression, respectively. In other words, these parameters set the strength of feedback on HilC and RtsA expression. In these simulations, the parameters
specify the degree by which the SPI1 regulators - HilC, HilD, and RtsA - can activate HilC and RtsA expression, respectively. In other words, these parameters set the strength of feedback on HilC and RtsA expression. In these simulations, the parameters  and
and  were both varied in tandem: the numerical values for the two are the same. (C) Effect of HilE on HilD expression. Plot shows the steady-state concentration of HilD as a function of the parameters
were both varied in tandem: the numerical values for the two are the same. (C) Effect of HilE on HilD expression. Plot shows the steady-state concentration of HilD as a function of the parameters  and
and  . The parameter
. The parameter  specifies the rate of HilE expression. Results for HilA are shown in Figures S6A–C. The black lines in the plots are used to denote the results obtained using the nominal parameters (aside from
specifies the rate of HilE expression. Results for HilA are shown in Figures S6A–C. The black lines in the plots are used to denote the results obtained using the nominal parameters (aside from  ). A detailed description of the model is provided in the Materials and Methods.
). A detailed description of the model is provided in the Materials and Methods.
Similar articles
-
Correction: The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella.PLoS Pathog. 2010 Aug 5;6(8):10.1371/annotation/df7e26bc-4c62-43b4-865f-a39274d98ab3. doi: 10.1371/annotation/df7e26bc-4c62-43b4-865f-a39274d98ab3. PLoS Pathog. 2010. PMID: 20700452 Free PMC article.
-
The Small RNA MicC Downregulates hilD Translation To Control the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2022 Jan 18;204(1):e0037821. doi: 10.1128/JB.00378-21. Epub 2021 Oct 25. J Bacteriol. 2022. PMID: 34694902 Free PMC article.
-
HilD, HilC, and RtsA Form Homodimers and Heterodimers To Regulate Expression of the Salmonella Pathogenicity Island I Type III Secretion System.J Bacteriol. 2020 Apr 9;202(9):e00012-20. doi: 10.1128/JB.00012-20. Print 2020 Apr 9. J Bacteriol. 2020. PMID: 32041797 Free PMC article.
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
-
The Salmonella pathogenicity island-1 type III secretion system.Microbes Infect. 2001 Nov-Dec;3(14-15):1281-91. doi: 10.1016/s1286-4579(01)01488-5. Microbes Infect. 2001. PMID: 11755416 Review.
Cited by
-
FliZ induces a kinetic switch in flagellar gene expression.J Bacteriol. 2010 Dec;192(24):6477-81. doi: 10.1128/JB.00751-10. Epub 2010 Oct 8. J Bacteriol. 2010. PMID: 20935096 Free PMC article.
-
For the Greater (Bacterial) Good: Heterogeneous Expression of Energetically Costly Virulence Factors.Infect Immun. 2020 Jun 22;88(7):e00911-19. doi: 10.1128/IAI.00911-19. Print 2020 Jun 22. Infect Immun. 2020. PMID: 32041785 Free PMC article. Review.
-
Long-Distance Effects of H-NS Binding in the Control of hilD Expression in the Salmonella SPI1 Locus.J Bacteriol. 2021 Oct 12;203(21):e0030821. doi: 10.1128/JB.00308-21. Epub 2021 Aug 23. J Bacteriol. 2021. PMID: 34424033 Free PMC article.
-
In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD.Sci Rep. 2016 Nov 25;6:37858. doi: 10.1038/srep37858. Sci Rep. 2016. PMID: 27886269 Free PMC article.
-
Pseudomonas syringae subpopulations cooperate by coordinating flagellar and type III secretion spatiotemporal dynamics to facilitate plant infection.Nat Microbiol. 2025 Apr;10(4):958-972. doi: 10.1038/s41564-025-01966-0. Epub 2025 Apr 2. Nat Microbiol. 2025. PMID: 40175722 Free PMC article.
References
-
- Ellermeier CD, Slauch JM. The genus Salmonella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes, 3rd ed. New York, NY.: Springer; 2006. pp. 123–158.
-
- Miller SI, Pegues PA. Salmonella species, including Salmonella typhi. In: Bennett JE, Dolin R, editors. Principles of infectious diseases. Philadelphia PA: Churchill Livingstone; 2000. pp. 2344–2363.
-
- Mills DM, Bajaj V, Lee CA. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

