A comprehensive, quantitative, and genome-wide model of translation
- PMID: 20686685
- PMCID: PMC2912337
- DOI: 10.1371/journal.pcbi.1000865
A comprehensive, quantitative, and genome-wide model of translation
Abstract
Translation is still poorly characterised at the level of individual proteins and its role in regulation of gene expression has been constantly underestimated. To better understand the process of protein synthesis we developed a comprehensive and quantitative model of translation, characterising protein synthesis separately for individual genes. The main advantage of the model is that basing it on only a few datasets and general assumptions allows the calculation of many important translational parameters, which are extremely difficult to measure experimentally. In the model, each gene is attributed with a set of translational parameters, namely the absolute number of transcripts, ribosome density, mean codon translation time, total transcript translation time, total time required for translation initiation and elongation, translation initiation rate, mean mRNA lifetime, and absolute number of proteins produced by gene transcripts. Most parameters were calculated based on only one experimental dataset of genome-wide ribosome profiling. The model was implemented in Saccharomyces cerevisiae, and its results were compared with available data, yielding reasonably good correlations. The calculated coefficients were used to perform a global analysis of translation in yeast, revealing some interesting aspects of the process. We have shown that two commonly used measures of translation efficiency - ribosome density and number of protein molecules produced - are affected by two distinct factors. High values of both measures are caused, i.a., by very short times of translation initiation, however, the origins of initiation time reduction are completely different in both cases. The model is universal and can be applied to any organism, if the necessary input data are available. The model allows us to better integrate transcriptomic and proteomic data. A few other possibilities of the model utilisation are discussed concerning the example of the yeast system.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
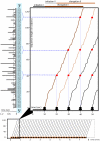
 (left) and
(left) and  (right) with experimentally determined mRNA and protein abundances by two independent studies , . The axes were log transformed. Calculated
(right) with experimentally determined mRNA and protein abundances by two independent studies , . The axes were log transformed. Calculated  values are presented in Table 2. The distribution of the log-fold differences of the mRNA and protein concentrations reported by the model and reference studies are presented in Supplementary Figure S1.
values are presented in Table 2. The distribution of the log-fold differences of the mRNA and protein concentrations reported by the model and reference studies are presented in Supplementary Figure S1.
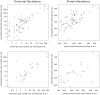
 with mRNA abundances determined by high-density oligonucleotide array (HDA) experiment . The axes were log transformed. Calculated
with mRNA abundances determined by high-density oligonucleotide array (HDA) experiment . The axes were log transformed. Calculated  value for the comparison is presented in Table 2. Right plot: distribution of the log-fold differences of the mRNA concentrations reported by the model and reference study.
value for the comparison is presented in Table 2. Right plot: distribution of the log-fold differences of the mRNA concentrations reported by the model and reference study.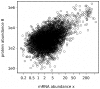
 ) and the number of protein molecules produced from a given gene (parameter
) and the number of protein molecules produced from a given gene (parameter  ). We performed linear regression through the origin on log transformed data. Adjusted
). We performed linear regression through the origin on log transformed data. Adjusted  value calculated over the entire dataset (4192 genes of known
value calculated over the entire dataset (4192 genes of known  ) was 0.59. This means that over 40% (in log space) of the variation in protein abundance cannot be explained by variation in mRNA abundance, suggesting some additional, posttranscriptional mechanisms of gene expression regulation.
) was 0.59. This means that over 40% (in log space) of the variation in protein abundance cannot be explained by variation in mRNA abundance, suggesting some additional, posttranscriptional mechanisms of gene expression regulation.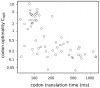
 C of individual yeast codons with codon optimality values
C of individual yeast codons with codon optimality values  calculated by . There is negative correlation between
calculated by . There is negative correlation between  value and translation time of a codon. However, while optimal codons (high
value and translation time of a codon. However, while optimal codons (high  values) have only short times of translation, non-optimal codons may be translated at both high and low rates. Adjusted
values) have only short times of translation, non-optimal codons may be translated at both high and low rates. Adjusted  value obtained in linear regression model through the origin on log transformed values indicates, that translation speed may explain only 15% of variability in
value obtained in linear regression model through the origin on log transformed values indicates, that translation speed may explain only 15% of variability in  values.
values.References
-
- Kochetov AV, Kolchanov NA, Sarai A. Interrelations between the efficiency of translation start sites and other sequence features of yeast mRNAs. Mol Genet Genomics. 2003;270:442–7. - PubMed
-
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–70. - PubMed
-
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–56. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

