More than 1,001 problems with protein domain databases: transmembrane regions, signal peptides and the issue of sequence homology
- PMID: 20686689
- PMCID: PMC2912341
- DOI: 10.1371/journal.pcbi.1000867
More than 1,001 problems with protein domain databases: transmembrane regions, signal peptides and the issue of sequence homology
Abstract
Large-scale genome sequencing gained general importance for life science because functional annotation of otherwise experimentally uncharacterized sequences is made possible by the theory of biomolecular sequence homology. Historically, the paradigm of similarity of protein sequences implying common structure, function and ancestry was generalized based on studies of globular domains. Having the same fold imposes strict conditions over the packing in the hydrophobic core requiring similarity of hydrophobic patterns. The implications of sequence similarity among non-globular protein segments have not been studied to the same extent; nevertheless, homology considerations are silently extended for them. This appears especially detrimental in the case of transmembrane helices (TMs) and signal peptides (SPs) where sequence similarity is necessarily a consequence of physical requirements rather than common ancestry. Thus, matching of SPs/TMs creates the illusion of matching hydrophobic cores. Therefore, inclusion of SPs/TMs into domain models can give rise to wrong annotations. More than 1001 domains among the 10,340 models of Pfam release 23 and 18 domains of SMART version 6 (out of 809) contain SP/TM regions. As expected, fragment-mode HMM searches generate promiscuous hits limited to solely the SP/TM part among clearly unrelated proteins. More worryingly, we show explicit examples that the scores of clearly false-positive hits, even in global-mode searches, can be elevated into the significance range just by matching the hydrophobic runs. In the PIR iProClass database v3.74 using conservative criteria, we find that at least between 2.1% and 13.6% of its annotated Pfam hits appear unjustified for a set of validated domain models. Thus, false-positive domain hits enforced by SP/TM regions can lead to dramatic annotation errors where the hit has nothing in common with the problematic domain model except the SP/TM region itself. We suggest a workflow of flagging problematic hits arising from SP/TM-containing models for critical reconsideration by annotation users.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
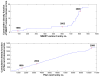
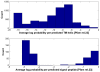
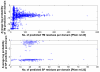
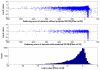
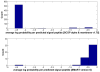
 is the database size,
is the database size,  and
and  are the extreme value distribution (EVD) parameters of the domain model. The bottom plot depicts the histogram of the 10340 domains in Pfam rel.23. The median of all log E-values that corresponded to the domain-specific GAs is found to be −1.16. This translates to an E-value of 0.07.
are the extreme value distribution (EVD) parameters of the domain model. The bottom plot depicts the histogram of the 10340 domains in Pfam rel.23. The median of all log E-values that corresponded to the domain-specific GAs is found to be −1.16. This translates to an E-value of 0.07.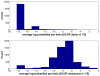
Similar articles
-
dissectHMMER: a HMMER-based score dissection framework that statistically evaluates fold-critical sequence segments for domain fold similarity.Biol Direct. 2015 Aug 1;10:39. doi: 10.1186/s13062-015-0068-3. Biol Direct. 2015. PMID: 26228544 Free PMC article.
-
Not all transmembrane helices are born equal: Towards the extension of the sequence homology concept to membrane proteins.Biol Direct. 2011 Oct 25;6:57. doi: 10.1186/1745-6150-6-57. Biol Direct. 2011. PMID: 22024092 Free PMC article.
-
xHMMER3x2: Utilizing HMMER3's speed and HMMER2's sensitivity and specificity in the glocal alignment mode for improved large-scale protein domain annotation.Biol Direct. 2016 Nov 29;11(1):63. doi: 10.1186/s13062-016-0163-0. Biol Direct. 2016. PMID: 27894340 Free PMC article.
-
Protein family classification and functional annotation.Comput Biol Chem. 2003 Feb;27(1):37-47. doi: 10.1016/s1476-9271(02)00098-1. Comput Biol Chem. 2003. PMID: 12798038 Review.
-
Computational analysis of protein tyrosine phosphatases: practical guide to bioinformatics and data resources.Methods. 2005 Jan;35(1):90-114. doi: 10.1016/j.ymeth.2004.07.012. Methods. 2005. PMID: 15588990 Review.
Cited by
-
dissectHMMER: a HMMER-based score dissection framework that statistically evaluates fold-critical sequence segments for domain fold similarity.Biol Direct. 2015 Aug 1;10:39. doi: 10.1186/s13062-015-0068-3. Biol Direct. 2015. PMID: 26228544 Free PMC article.
-
fLPS 2.0: rapid annotation of compositionally-biased regions in biological sequences.PeerJ. 2021 Oct 28;9:e12363. doi: 10.7717/peerj.12363. eCollection 2021. PeerJ. 2021. PMID: 34760378 Free PMC article.
-
ULTRA: A Model Based Tool to Detect Tandem Repeats.ACM BCB. 2018 Aug-Sep;2018:37-46. doi: 10.1145/3233547.3233604. ACM BCB. 2018. PMID: 31080962 Free PMC article.
-
Sequence-divergent chordopoxvirus homologs of the o3 protein maintain functional interactions with components of the vaccinia virus entry-fusion complex.J Virol. 2012 Feb;86(3):1696-705. doi: 10.1128/JVI.06069-11. Epub 2011 Nov 23. J Virol. 2012. PMID: 22114343 Free PMC article.
-
Comparative genomics of transport proteins in seven Bacteroides species.PLoS One. 2018 Dec 5;13(12):e0208151. doi: 10.1371/journal.pone.0208151. eCollection 2018. PLoS One. 2018. PMID: 30517169 Free PMC article.
References
-
- Eisenhaber F. Prediction of Protein Function: Two Basic Concepts and One Practical Recipe. In: Eisenhaber F, editor. Discovering Biomolecular Mechanisms with Computational Biology. Georgetown and New York: Landes Biosciences and Springer; 2006. pp. 39–54.
-
- Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9:210–219. - PubMed
-
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, et al. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. - PubMed
-
- Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, et al. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–725. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

