Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1
- PMID: 20687133
- PMCID: PMC2998726
- DOI: 10.1002/pro.479
Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1
Abstract
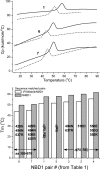
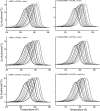
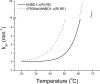
Misfolding and degradation of CFTR is the cause of disease in patients with the most prevalent CFTR mutation, an in-frame deletion of phenylalanine (F508del), located in the first nucleotide-binding domain of human CFTR (hNBD1). Studies of (F508del)CFTR cellular folding suggest that both intra- and inter-domain folding is impaired. (F508del)CFTR is a temperature-sensitive mutant, that is, lowering growth temperature, improves both export, and plasma membrane residence times. Yet, paradoxically, F508del does not alter the fold of isolated hNBD1 nor did it seem to perturb its unfolding transition in previous isothermal chemical denaturation studies. We therefore studied the in vitro thermal unfolding of matched hNBD1 constructs ±F508del to shed light on the defective folding mechanism and the basis for the thermal instability of (F508del)CFTR. Using primarily differential scanning calorimetry (DSC) and circular dichroism, we show for all hNBD1 pairs studied, that F508del lowers the unfolding transition temperature (T(m)) by 6-7°C and that unfolding occurs via a kinetically-controlled, irreversible transition in isolated monomers. A thermal unfolding mechanism is derived from nonlinear least squares fitting of comprehensive DSC data sets. All data are consistent with a simple three-state thermal unfolding mechanism for hNBD1 ± F508del: N(±MgATP) <==> I(T)(±MgATP) → A(T) → (A(T))(n). The equilibrium unfolding to intermediate, I(T), is followed by the rate-determining, irreversible formation of a partially folded, aggregation-prone, monomeric state, A(T), for which aggregation to (A(T))(n) and further unfolding occur with no detectable heat change. Fitted parameters indicate that F508del thermodynamically destabilizes the native state, N, and accelerates the formation of A(T).
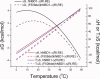
Figures


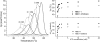
References
-
- Zielenski J, Tsui LC. Cystic fibrosis: genotypic and phenotypic variations. Annu Rev Genet. 1995;29:777–807. - PubMed
-
- Riordan JR. CFTR function and prospects for therapy. Annu Rev Biochem. 2008;77:701–726. - PubMed
-
- Riordan JR. Assembly of functional CFTR chloride channels. Annu Rev Physiol. 2005;67:701–718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

