RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome
- PMID: 20687943
- PMCID: PMC3017786
- DOI: 10.1186/1471-2229-10-160
RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome
Abstract
Background: Next generation sequencing is transforming our understanding of transcriptomes. It can determine the expression level of transcripts with a dynamic range of over six orders of magnitude from multiple tissues, developmental stages or conditions. Patterns of gene expression provide insight into functions of genes with unknown annotation.
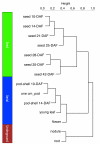
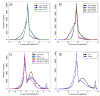
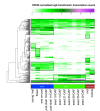
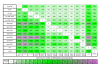
Results: The RNA Seq-Atlas presented here provides a record of high-resolution gene expression in a set of fourteen diverse tissues. Hierarchical clustering of transcriptional profiles for these tissues suggests three clades with similar profiles: aerial, underground and seed tissues. We also investigate the relationship between gene structure and gene expression and find a correlation between gene length and expression. Additionally, we find dramatic tissue-specific gene expression of both the most highly-expressed genes and the genes specific to legumes in seed development and nodule tissues. Analysis of the gene expression profiles of over 2,000 genes with preferential gene expression in seed suggests there are more than 177 genes with functional roles that are involved in the economically important seed filling process. Finally, the Seq-atlas also provides a means of evaluating existing gene model annotations for the Glycine max genome.
Conclusions: This RNA-Seq atlas extends the analyses of previous gene expression atlases performed using Affymetrix GeneChip technology and provides an example of new methods to accommodate the increase in transcriptome data obtained from next generation sequencing. Data contained within this RNA-Seq atlas of Glycine max can be explored at http://www.soybase.org/soyseq.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

