BST-2/tetherin: a new component of the innate immune response to enveloped viruses
- PMID: 20688520
- PMCID: PMC2956607
- DOI: 10.1016/j.tim.2010.06.010
BST-2/tetherin: a new component of the innate immune response to enveloped viruses
Abstract
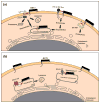
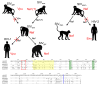
The interferon-inducible, transmembrane protein BST-2 (CD317, tetherin) directly holds fully formed enveloped virus particles to the cells that produce them, inhibiting their spread. BST-2 inhibits members of the retrovirus, filovirus, arenavirus and herpesvirus families. These viruses encode a variety of proteins to degrade BST-2 and/or direct it away from its site of action at the cell surface. Viral antagonism has subjected BST-2 to positive selection, leading to species-specific differences that presented a barrier to the transmission of simian immunodeficiency viruses (SIVs) to humans. This barrier was crossed by HIV-1 when its Vpu protein acquired activity as a BST-2 antagonist. Here, we review this new host-pathogen relationship and discuss its impact on the evolution of primate lentiviruses and the origins of the HIV pandemic.
Published by Elsevier Ltd.
Figures

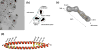
References
-
- Neil SJ, et al. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. - PubMed
-
- Ishikawa J, et al. Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST2, that may be involved in pre-B-cell growth. Genomics. 1995;26:527–534. - PubMed
-
- Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
-
- Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

