Mycobacterium marinum MMAR_2380, a predicted transmembrane acyltransferase, is essential for the presence of the mannose cap on lipoarabinomannan
- PMID: 20688818
- PMCID: PMC3090144
- DOI: 10.1099/mic.0.037507-0
Mycobacterium marinum MMAR_2380, a predicted transmembrane acyltransferase, is essential for the presence of the mannose cap on lipoarabinomannan
Abstract
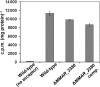
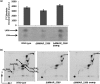
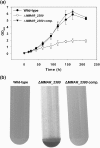
Lipoarabinomannan (LAM) is a major glycolipid in the mycobacterial cell envelope. LAM consists of a mannosylphosphatidylinositol (MPI) anchor, a mannan core and a branched arabinan domain. The termini of the arabinan branches can become substituted with one to three α(1→2)-linked mannosyl residues, the mannose cap, producing ManLAM. ManLAM has been associated with a range of different immunomodulatory properties of Mycobacterium tuberculosis during infection of the host. In some of these effects, the presence of the mannose cap on ManLAM appears to be crucial for its activity. So far, in the biosynthesis of the mannose cap on ManLAM, two enzymes have been reported to be involved: a mannosyltransferase that adds the first mannosyl residue of the mannose caps to the arabinan domain of LAM, and another mannosyltransferase that elongates the mannose cap up to three mannosyl residues. Here, we report that a third gene is involved, MMAR_2380, which is the Mycobacterium marinum orthologue of Rv1565c. MMAR_2380 encodes a predicted transmembrane acyltransferase. In M. marinum ΔMMAR_2380, the LAM arabinan domain is still intact, but the mutant LAM lacks the mannose cap. Additional effects of mutation of MMAR_2380 on LAM were observed: a higher degree of branching of both the arabinan domain and the mannan core, and a decreased incorporation of [1,2-(14)C]acetate into the acyl chains in mutant LAM as compared with the wild-type form. This latter effect was also observed for related lipoglycans, i.e. lipomannan (LM) and phosphatidylinositol mannosides (PIMs). Furthermore, the mutant strain showed increased aggregation in liquid cultures as compared with the wild-type strain. All phenotypic traits of M. marinum ΔMMAR_2380, the deficiency in the mannose cap on LAM and changes at the cell surface, could be reversed by complementing the mutant strain with MMAR_2380. Strikingly, membrane preparations of the mutant strain still showed enzymic activity for the arabinan mannose-capping mannosyltransferase similar to that of the wild-type strain. Although the exact function of MMAR_2380 remains unknown, we show that the protein is essential for the presence of a mannose cap on LAM.
Figures

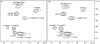
Similar articles
-
Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase.Proc Natl Acad Sci U S A. 2008 Nov 18;105(46):17973-7. doi: 10.1073/pnas.0807761105. Epub 2008 Nov 12. Proc Natl Acad Sci U S A. 2008. PMID: 19004785 Free PMC article.
-
A single arabinan chain is attached to the phosphatidylinositol mannosyl core of the major immunomodulatory mycobacterial cell envelope glycoconjugate, lipoarabinomannan.J Biol Chem. 2014 Oct 31;289(44):30249-30256. doi: 10.1074/jbc.M114.599415. Epub 2014 Sep 17. J Biol Chem. 2014. PMID: 25231986 Free PMC article.
-
Mannan core branching of lipo(arabino)mannan is required for mycobacterial virulence in the context of innate immunity.Cell Microbiol. 2013 Dec;15(12):2093-108. doi: 10.1111/cmi.12175. Epub 2013 Aug 22. Cell Microbiol. 2013. PMID: 23902464 Free PMC article.
-
Mannose-capped lipoarabinomannan in Mycobacterium tuberculosis pathogenesis.Pathog Dis. 2018 Jun 1;76(4):fty026. doi: 10.1093/femspd/fty026. Pathog Dis. 2018. PMID: 29722821 Free PMC article. Review.
-
Lipoarabinomannan, and its related glycolipids, induce divergent and opposing immune responses to Mycobacterium tuberculosis depending on structural diversity and experimental variations.Tuberculosis (Edinb). 2016 Jan;96:120-30. doi: 10.1016/j.tube.2015.09.005. Epub 2015 Oct 28. Tuberculosis (Edinb). 2016. PMID: 26586646 Review.
Cited by
-
Role of succinyl substituents in the mannose-capping of lipoarabinomannan and control of inflammation in Mycobacterium tuberculosis infection.PLoS Pathog. 2023 Sep 5;19(9):e1011636. doi: 10.1371/journal.ppat.1011636. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37669276 Free PMC article.
-
Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction.FEMS Microbiol Rev. 2011 Nov;35(6):1126-57. doi: 10.1111/j.1574-6976.2011.00276.x. Epub 2011 May 31. FEMS Microbiol Rev. 2011. PMID: 21521247 Free PMC article. Review.
-
Transposon mutagenesis in Mycobacterium kansasii links a small RNA gene to colony morphology and biofilm formation and identifies 9,885 intragenic insertions that do not compromise colony outgrowth.Microbiologyopen. 2020 Apr;9(4):e988. doi: 10.1002/mbo3.988. Epub 2020 Feb 21. Microbiologyopen. 2020. PMID: 32083796 Free PMC article.
-
Lipoarabinomannan modification as a source of phenotypic heterogeneity in host-adapted Mycobacterium abscessus isolates.Proc Natl Acad Sci U S A. 2024 Apr 23;121(17):e2403206121. doi: 10.1073/pnas.2403206121. Epub 2024 Apr 17. Proc Natl Acad Sci U S A. 2024. PMID: 38630725 Free PMC article.
-
Synthetic lethality reveals mechanisms of Mycobacterium tuberculosis resistance to β-lactams.mBio. 2014 Sep 16;5(5):e01767-14. doi: 10.1128/mBio.01767-14. mBio. 2014. PMID: 25227469 Free PMC article.
References
-
- Abdallah, A. M., Verboom, T., Hannes, F., Safi, M., Strong, M., Eisenberg, D., Musters, R. J., Vandenbroucke-Grauls, C. M., Appelmelk, B. J. & other authors (2006). A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol 62, 667–679. - PubMed
-
- Appelmelk, B. J., den Dunnen, J., Driessen, N. N., Ummels, R., Pak, M., Nigou, J., Larrouy-Maumus, G., Gurcha, S. S., Movahedzadeh, F. & other authors (2008). The mannose cap of mycobacterial lipoarabinomannan does not dominate the Mycobacterium–host interaction. Cell Microbiol 10, 930–944. - PubMed
-
- Belisle, J. T., Klaczkiewicz, K., Brennan, P. J., Jacobs, W. R. & Inamine, J. M. (1993). Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J Biol Chem 268, 10517–10523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

