A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R
- PMID: 20688916
- PMCID: PMC2937936
- DOI: 10.1074/jbc.C110.168641
A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R
Abstract
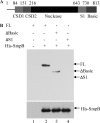
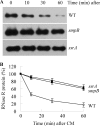
The amount of RNase R, an important degradative exoribonuclease, increases 3-10-fold under a variety of stress conditions. This elevation is due to posttranslational regulation in which the highly unstable RNase R protein becomes stabilized during stress. Here we identify two components of the trans-translation machinery, transfer-messenger RNA (tmRNA) and SmpB, that are responsible for the short half-life of RNase R in exponential phase cells. The absence of either lengthens the half-life of RNase R in vivo >6-fold. SmpB directly interacts with RNase R in vitro and is stimulated by tmRNA. The C-terminal region of RNase R, encompassing its basic region and adjacent S1 domain are required for the interaction; their removal eliminates binding and stabilizes RNase R in vivo. However, the binding of SmpB and tmRNA does not alter RNase R activity. These data define a previously unknown regulatory process in which the stability of an RNase is determined by its interaction with an RNA and an RNA-associated protein.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

